1IWT
 
 | | Crystal Structure Analysis of Human lysozyme at 113K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4EB1
 
 | | Hyperstable in-frame insertion variant of antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III | | Authors: | Martinez-Martinez, I, Johnson, D.J.D, Yamasaki, M, Corral, J, Huntington, J.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Type II antithrombin deficiency caused by a large in-frame insertion: structural, functional and pathological relevance.
J.Thromb.Haemost., 10, 2012
|
|
1IWX
 
 | | Crystal Structure Analysis of Human lysozyme at 161K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1Y3H
 
 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD kinase from Mycobacterium tuberculosis | | Descriptor: | Inorganic polyphosphate/ATP-NAD kinase | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
BIOCHEM.BIOPHYS.RES.COMMUN., 327, 2005
|
|
1JWR
 
 | | Crystal structure of human lysozyme at 100 K | | Descriptor: | lysozyme | | Authors: | Higo, J, Nakasako, M. | | Deposit date: | 2001-09-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydration structure of human lysozyme investigated by molecular dynamics simulation and cryogenic X-ray crystal structure analyses: on the correlation between crystal water sites, solvent density, and solvent dipole
J.Comput.Chem., 23, 2002
|
|
1MHU
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
3VTV
 
 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | Descriptor: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAO
 
 | | Crystal structure of Atg13 LIR-fused human LC3B_2-119 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAN
 
 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAM
 
 | | Crystal structure of human LC3C_8-125 | | Descriptor: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
1TH5
 
 | | Solution structure of C-terminal domain of NifU-like protein from Oryza sativa | | Descriptor: | NifU1 | | Authors: | Kumeta, H, Ogura, K, Asayama, M, Katoh, S, Katoh, E, Inagaki, F. | | Deposit date: | 2004-06-01 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the domain II of a chloroplastic NifU-like protein OsNifU1A.
J.Biomol.Nmr, 38, 2007
|
|
1SXB
 
 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
1SU4
 
 | | Crystal structure of calcium ATPase with two bound calcium ions | | Descriptor: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Toyoshima, C, Nakasako, M, Nomura, H, Ogawa, H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution
Nature, 405, 2000
|
|
1SXA
 
 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
1SXC
 
 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
1VFI
 
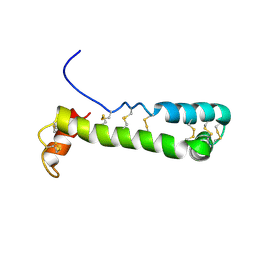 | | Solution Structure of Vanabin2 (RUH-017), a Vanadium-binding Protein from Ascidia sydneiensis samea | | Descriptor: | vanadium-binding protein 2 | | Authors: | Hamada, T, Hirota, H, Asanuma, M, Hayashi, F, Kobayashi, N, Ueki, T, Michibata, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vanabin2, a Vanadium(IV)-Binding Protein from the Vanadium-Rich Ascidian Ascidia sydneiensis samea
J.Am.Chem.Soc., 127, 2005
|
|
2ZAB
 
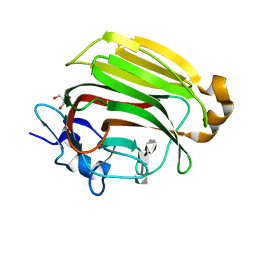 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Cmplex with Product (GGG) | | Descriptor: | Alginate lyase, GLYCEROL, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZGY
 
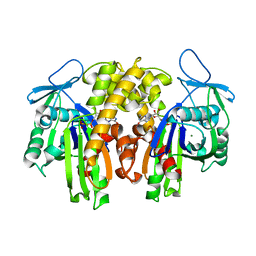 | | PARM with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
2ZAC
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Complex with Product (MMG) | | Descriptor: | Alginate lyase, GLYCEROL, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZHC
 
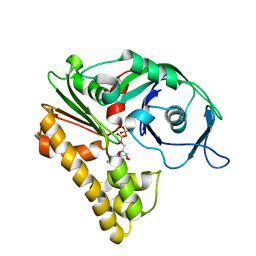 | | ParM filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
3A0N
 
 | | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
3BIK
 
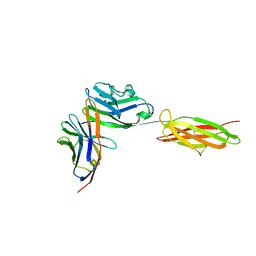 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZA9
 
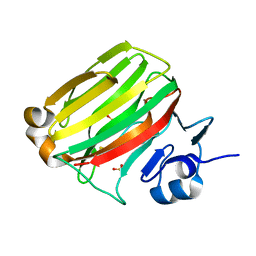 | | Crystal Structure of Alginate lyase A1-II' N141C/N199C | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
