6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | 分子名称: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | 著者 | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | 登録日 | 2019-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
6HZN
 
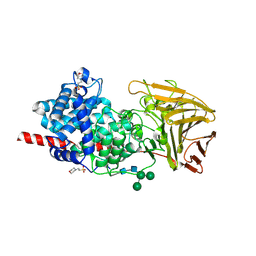 | | Crystal structure of human dermatan sulfate epimerase 1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hasan, M, Unge, J, Westergren-Thorsson, G, Ellervik, U, Mueller, U, Malmstrom, A, Tykesson, E. | | 登録日 | 2018-10-23 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | The structure of human dermatan sulfate epimerase 1 emphasizes the importance of C5-epimerization of glucuronic acid in higher organisms
Chem Sci, 2020
|
|
6L96
 
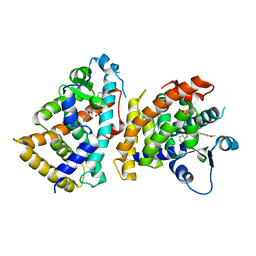 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | 著者 | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | 登録日 | 2019-11-08 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
6A5I
 
 | |
3EIS
 
 | |
3DTV
 
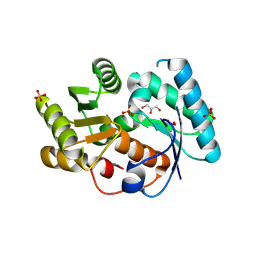 | | Crystal structure of arylmalonate decarboxylase | | 分子名称: | Arylmalonate decarboxylase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | 著者 | Nakasako, M, Obata, R, Miyamaoto, K, Ohta, H. | | 登録日 | 2008-07-16 | | 公開日 | 2009-07-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Inverting the Enantioselectivity of Arylmalonate Decarboxylase Revealed by the Structural Analysis of the Gly74Cys/Cys188Ser Mutant in the Liganded Form
Biochemistry, 49, 2010
|
|
3IXL
 
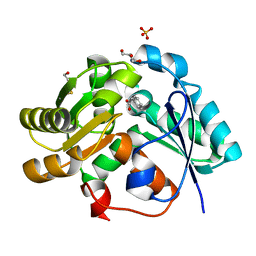 | |
3IXM
 
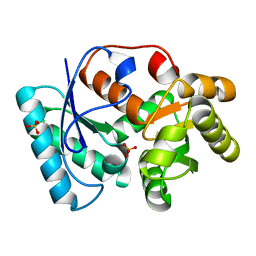 | |
1WR6
 
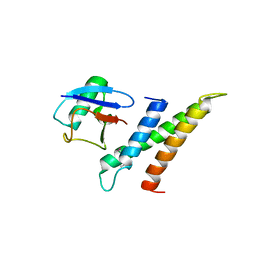 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | 分子名称: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | 著者 | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | 登録日 | 2004-10-12 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
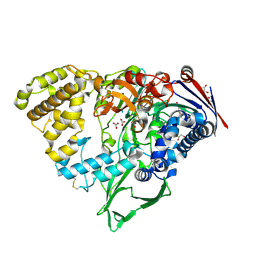
7EBY
 
 | |
7EA4
 
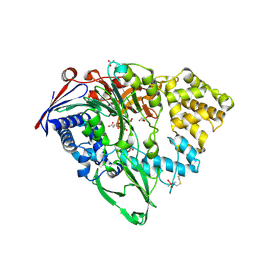 | |
2STD
 
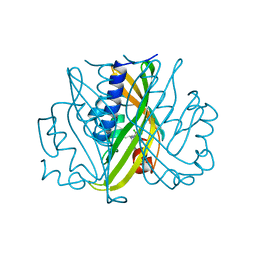 | | SCYTALONE DEHYDRATASE COMPLEXED WITH TIGHT-BINDING INHIBITOR CARPROPAMID | | 分子名称: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, SCYTALONE DEHYDRATASE, SULFATE ION | | 著者 | Nakasako, M, Motoyama, T, Kurahashi, Y, Yamaguchi, I. | | 登録日 | 1997-12-21 | | 公開日 | 1999-02-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cryogenic X-ray crystal structure analysis for the complex of scytalone dehydratase of a rice blast fungus and its tight-binding inhibitor, carpropamid: the structural basis of tight-binding inhibition.
Biochemistry, 37, 1998
|
|
1DU1
 
 | |
1IDP
 
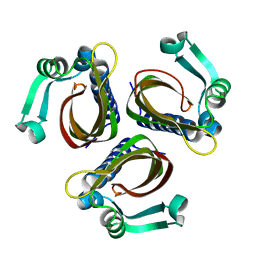 | |
1JTI
 
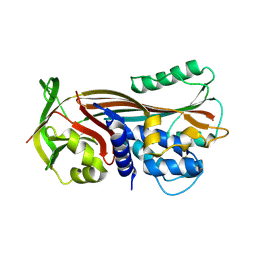 | |
1DLF
 
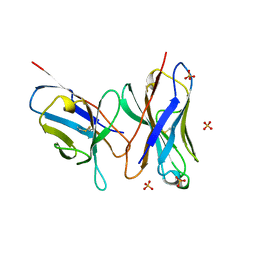 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | 分子名称: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | 著者 | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | 登録日 | 1998-07-14 | | 公開日 | 1999-07-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
1UHG
 
 | | Crystal Structure of S-Ovalbumin At 1.9 Angstrom Resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ovalbumin, ... | | 著者 | Yamasaki, M, Takahashi, N, Hirose, M. | | 登録日 | 2003-07-03 | | 公開日 | 2003-07-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of S-ovalbumin as a Non-loop-inserted Thermostabilized Serpin Form
J.Biol.Chem., 278, 2003
|
|
1WZ1
 
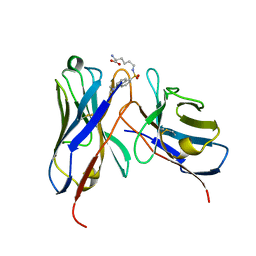 | | Crystal structure of the Fv fragment complexed with dansyl-lysine | | 分子名称: | Ig heavy chain, Ig light chain, N~6~-{[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}-L-LYSINE | | 著者 | Nakasako, M, Oka, T, Mashumo, M, Takahashi, H, Shimada, I, Yamaguchi, Y, Kato, K, Arata, Y. | | 登録日 | 2005-02-21 | | 公開日 | 2006-01-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conformational dynamics of complementarity-determining region H3 of an anti-dansyl Fv fragment in the presence of its hapten
J.Mol.Biol., 351, 2005
|
|
1EUZ
 
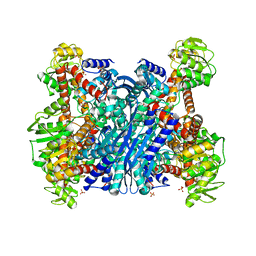 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS PROFUNDUS IN THE UNLIGATED STATE | | 分子名称: | GLUTAMATE DEHYDROGENASE, SULFATE ION | | 著者 | Nakasako, M. | | 登録日 | 2000-04-19 | | 公開日 | 2001-04-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Large-scale domain movements and hydration structure changes in the active-site cleft of unligated glutamate dehydrogenase from Thermococcus profundus studied by cryogenic X-ray crystal structure analysis and small-angle X-ray scattering.
Biochemistry, 40, 2001
|
|
1IO6
 
 | |
1VAV
 
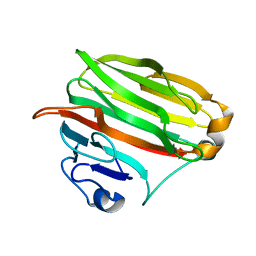 | | Crystal structure of alginate lyase PA1167 from Pseudomonas aeruginosa at 2.0 A resolution | | 分子名称: | Alginate lyase PA1167 | | 著者 | Yamasaki, M, Moriwaki, S, Miyake, O, Hashimoto, W, Murata, K, Mikami, B. | | 登録日 | 2004-02-19 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold.
J.Biol.Chem., 279, 2004
|
|
2DLF
 
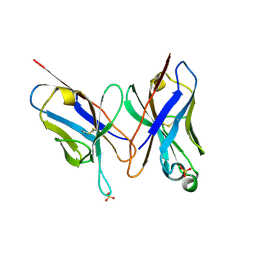 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | 分子名称: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | 著者 | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | 登録日 | 1998-12-17 | | 公開日 | 1999-12-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2Z6D
 
 | |
2Z6C
 
 | |
2CWS
 
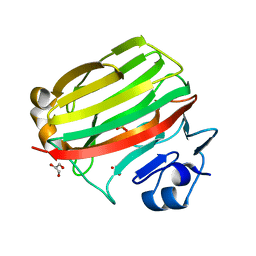 | | Crystal structure at 1.0 A of alginate lyase A1-II', a member of polysaccharide lyase family-7 | | 分子名称: | GLYCEROL, SULFATE ION, alginate lyase A1-II' | | 著者 | Yamasaki, M, Ogura, K, Hashimoto, W, Mikami, B, Murata, K. | | 登録日 | 2005-06-25 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | A Structural Basis for Depolymerization of Alginate by Polysaccharide Lyase Family-7
J.Mol.Biol., 352, 2005
|
|
