4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
6VFO
 
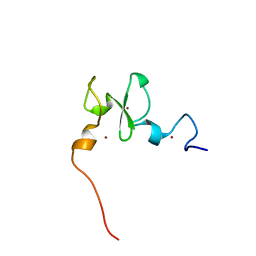 | | Solution structure of the PHD of mouse UHRF1 (NP95) | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Lemak, A, Houliston, S, Duan, S, Arrowsmith, C.H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
6VEE
 
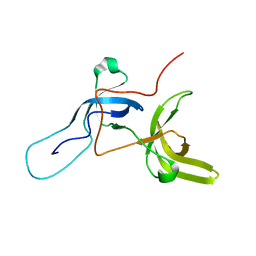 | |
6VED
 
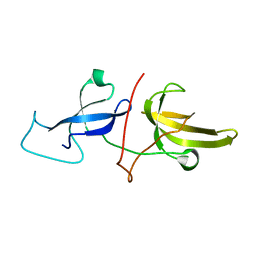 | | Solution structure of the TTD and linker region of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Lemak, A, Houliston, S, Duan, S, Ong, M.S, Arrowsmith, C.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
6VTI
 
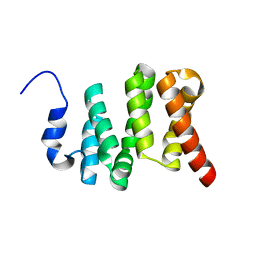 | | Solution NMR structure of the N-terminal domain of the Serine/threonine-protein phosphatase 1 regulatory subunit 10, PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2020-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
4XCX
 
 | | METHYLTRANSFERASE DOMAIN OF SMALL RNA 2'-O-METHYLTRANSFERASE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Small RNA 2'-O-methyltransferase | | Authors: | Walker, J.R, Zeng, H, Dong, A, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of human C1ORF59 in complex with SAH
To be published
|
|
6VWB
 
 | | Solution structure of the N-terminal helix-hairpin-helix domain of human MUS81 | | Descriptor: | Crossover junction endonuclease MUS81 | | Authors: | Payliss, B, Houliston, S, Lemak, A, Arrowsmith, C.H, Wyatt, H.D.M. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease
Cell Rep, 41, 2022
|
|
4Y03
 
 | | Crystal Structure of the fifth bromodomain of human PB1 in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, CITRIC ACID, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of the fifth bromodomain of human PB1 in complex with salicylic acid
To Be Published
|
|
1COK
 
 | |
8G45
 
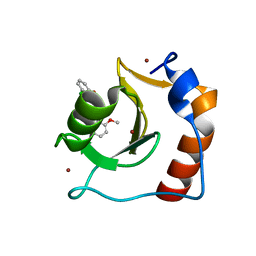 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G43
 
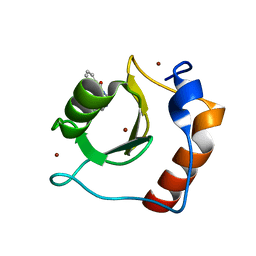 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G44
 
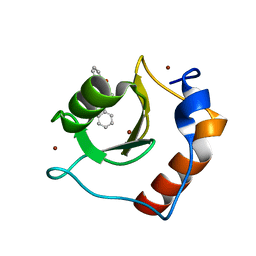 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
1HYW
 
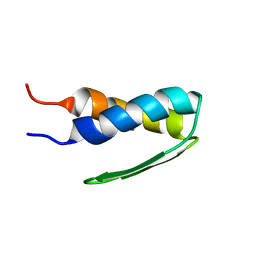 | | SOLUTION STRUCTURE OF BACTERIOPHAGE LAMBDA GPW | | Descriptor: | HEAD-TO-TAIL JOINING PROTEIN W | | Authors: | Maxwell, K.L, Yee, A.A, Booth, V, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold.
J.Mol.Biol., 308, 2001
|
|
1P9Q
 
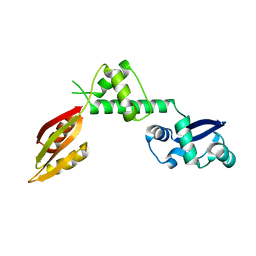 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1M94
 
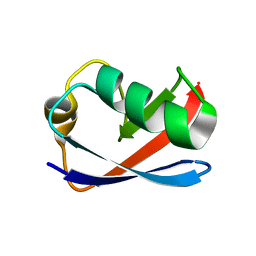 | | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1 | | Descriptor: | Protein YNR032c-a | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-07-26 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
6UPT
 
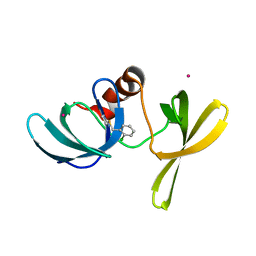 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706 | | Descriptor: | 2-((2-chlorobenzyl)thio)-4,5-dihydro-1H-imidazole, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706
to be published
|
|
1N6Z
 
 | |
6VAH
 
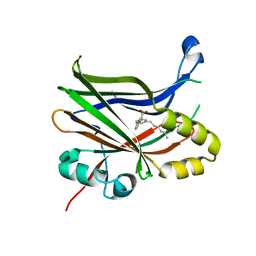 | | Crystal structure of human TEAD2 transcription factor in complex with Flufenamic acid derivative | | Descriptor: | 2-fluoro-6-[(3-hexylphenyl)amino]benzoic acid, Transcriptional enhancer factor TEF-4, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Dong, A, Li, Y, Melin, L, Gagnon, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human TEAD2 transcription factor in complex with Flufenamic acid derivative
to be published
|
|
6VA5
 
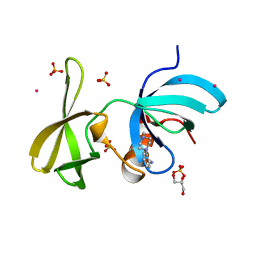 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-4184 | | Descriptor: | 2-(4-methylpiperazin-1-yl)aniline, GLYCEROL, SULFATE ION, ... | | Authors: | Zeng, H, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-4184
to be published
|
|
6VCS
 
 | | SRA domain of UHRF1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*AP*CP*AP*GP*GP*C)-3'), E3 ubiquitin-protein ligase UHRF1, UNK-UNK-UNK-UNK, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SRA domain of UHRF1 in complex with DNA
To Be Published
|
|
6VAN
 
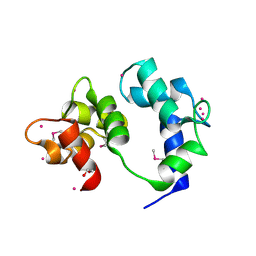 | | Crystal structure of caltubin from the great pond snail | | Descriptor: | 1,2-ETHANEDIOL, Caltubin, EF-hand, ... | | Authors: | Dong, A, Li, A, Zhang, Q, Barszczyk, A, Chern, Y.H, Arrowsmith, C.H, Edwards, A.M, Zhong, Z.P, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Cell-penetrating caltubin promotes neurite outgrowth and regrowth through calcium-dependent microtubule regulation
to be published
|
|
1IIO
 
 | |
1MV3
 
 | |
1MUZ
 
 | |
6VO5
 
 | | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4 | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4, ... | | Authors: | Halabelian, L, Zeng, H, Dong, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4
to be published
|
|
