5A3A
 
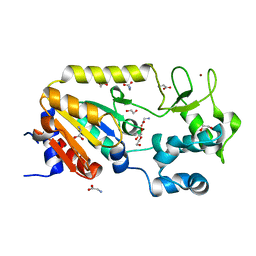 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
7OMY
 
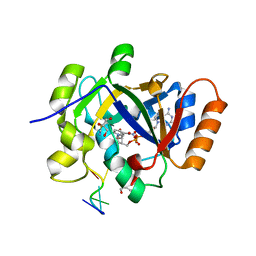 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ and ssDNA | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMX
 
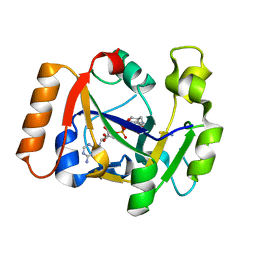 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DarT domain-containing protein, THIOCYANATE ION | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMV
 
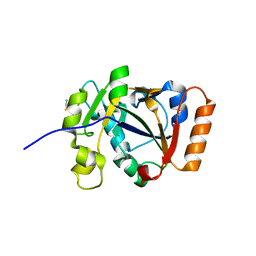 | | Thermus sp. 2.9 DarT | | Descriptor: | CHLORIDE ION, DarT domain-containing protein, THIOCYANATE ION | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMZ
 
 | |
7ON0
 
 | | Thermus sp. 2.9 DarT in complex with ADP-ribosylated ssDNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMW
 
 | | Thermus sp. 2.9 DarT in complex with NAD+ | | Descriptor: | DarT domain-containing protein, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
5A97
 
 | | Hazara virus nucleocapsid protain | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Surtees, R, Ariza, A, Hewson, R, Barr, J.N, Edwards, T.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Hazara Virus Nucleocapsid Protein.
Bmc Struct.Biol., 15, 2015
|
|
3ZHC
 
 | | Structure of the phytase from Citrobacter braakii at 2.3 angstrom resolution. | | Descriptor: | CHLORIDE ION, FORMIC ACID, PHYTASE | | Authors: | Wilson, K.S, Ariza, A, Sanchez-Romero, I, Skjot, M, Vind, J, DeMaria, L, Skov, L.K, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks
Plos One, 8, 2013
|
|
5A7R
 
 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
2WBA
 
 | | Properties of Trypanothione Reductase From T. brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Jones, D, Ariza, A, Chow, W.H.A, Oza, S.L, Fairlamb, A.H. | | Deposit date: | 2009-02-24 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative Structural, Kinetic and Inhibitor Studies of Trypanosoma Brucei Trypanothione Reductase with T. Cruzi.
Mol.Biochem.Parasitol., 169, 2010
|
|
2X8J
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | GLYCEROL, INTRACELLULAR SUBTILISIN PROTEASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Vedodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an intracellular subtilisin reveals novel structural features unique to this subtilisin family.
Structure, 18, 2010
|
|
2WWT
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | INTRACELLULAR SUBTILISIN PROTEASE, SODIUM ION | | Authors: | Vevodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2009-10-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of an Intracellular Subtilisin Reveals Novel Structural Features Unique to This Subtilisin Family.
Structure, 18, 2010
|
|
2WV7
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | INTRACELLULAR SUBTILISIN PROTEASE, SODIUM ION | | Authors: | Vevodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2009-10-15 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of an Intracellular Subtilisin Reveals Novel Structural Features Unique to This Subtilisin Family.
Structure, 18, 2010
|
|
8BAR
 
 | |
8BAS
 
 | | E. coli C7 DarT1 in complex with carba-NAD and DNA | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*AP*GP*AP*C)-3'), ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAT
 
 | | Geobacter lovleyi NADAR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Geobacter lovleyi NADAR | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAQ
 
 | | E. coli C7 DarT1 in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DarT ssDNA thymidine ADP-ribosyltransferase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAU
 
 | | Phytophthora nicotianae var. parasitica NADAR in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, NADAR domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
6SAO
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAV
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAU
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6FHV
 
 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FHW
 
 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FRV
 
 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
