8CKB
 
 | | Asymmetric reconstruction of the crAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Cargo protein 1 gp45, Head fiber dimer protein gp29, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural atlas of the most abundant human gut virus
Nature
|
|
9FFH
 
 | | Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
5OKF
 
 | | CH1 chimera of human 14-3-3 sigma with the HSPB6 phosphopeptide in a conformation with self-bound phosphopeptides | | Descriptor: | 14-3-3 protein sigma,Heat shock protein beta-6, CADMIUM ION | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
5OM0
 
 | | CH2 chimera of human 14-3-3 sigma with the Gli1 phosphopeptide around Ser640 | | Descriptor: | 14-3-3 protein sigma,Zinc finger protein GLI1, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
5OMA
 
 | | CH3 chimera of human 14-3-3 sigma with the StARD1 peptide including Ser57 | | Descriptor: | 14-3-3 protein sigma,Steroidogenic acute regulatory protein, mitochondrial, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
8CFA
 
 | |
8CEZ
 
 | |
4XVN
 
 | |
1QAW
 
 | | Regulatory Features of the TRP Operon and the Crystal Structure of the TRP RNA-Binding Attenuation Protein from Bacillus Stearothermophilus. | | Descriptor: | TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Chen, X.-P, Antson, A.A, Yang, M, Baumann, C, Dodson, E.J, Dodson, G.G, Gollnick, P. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus.
J.Mol.Biol., 289, 1999
|
|
7QIK
 
 | | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Smith, J.L.R, Antson, A.A. | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma
To Be Published
|
|
7QIP
 
 | |
3ZTE
 
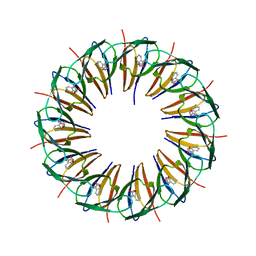 | |
4XP7
 
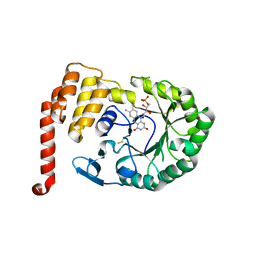 | | Crystal structure of Human tRNA dihydrouridine synthase 2 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Whelan, F, Jenkins, H.T, Griffiths, S, Byrne, R.T, Dodson, E.J, Antson, A.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From bacterial to human dihydrouridine synthase: automated structure determination.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GWE
 
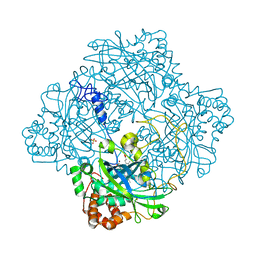 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GWH
 
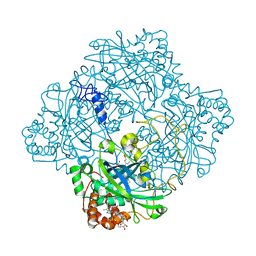 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase complexed with NADPH | | Descriptor: | ACETATE ION, Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4IWN
 
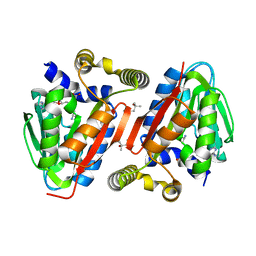 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | Authors: | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZQN
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 2) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQO
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 3) | | Descriptor: | POTASSIUM ION, TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQP
 
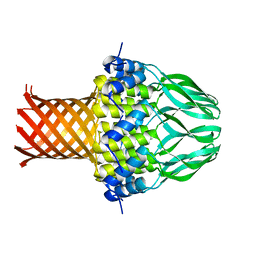 | | Crystal structure of the small terminase oligomerization domain from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQQ
 
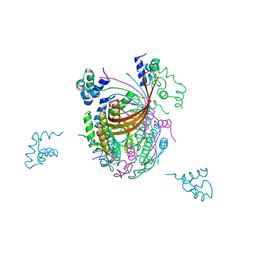 | | Crystal structure of the full-length small terminase from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZZL
 
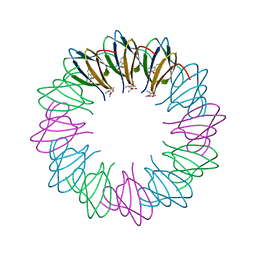 | | Bacillus halodurans trp RNA-binding attenuation protein (TRAP): a 12- subunit assembly | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZQM
 
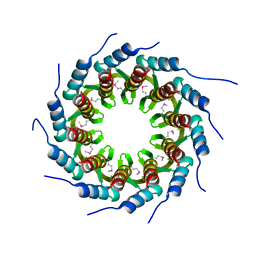 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1ODB
 
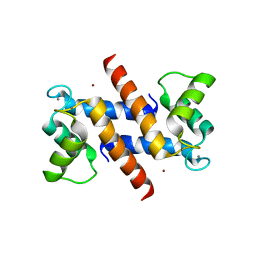 | | THE CRYSTAL STRUCTURE OF HUMAN S100A12 - COPPER COMPLEX | | Descriptor: | CALCIUM ION, CALGRANULIN C, COPPER (II) ION | | Authors: | Moroz, O.V, Antson, A.A, Grist, S.J, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2003-02-15 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7NT5
 
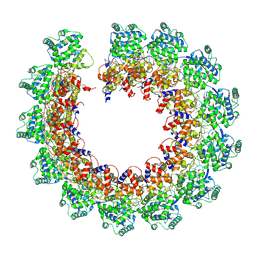 | | CryoEM structure of the Nipah virus nucleocapsid single helical turn assembly | | Descriptor: | Nucleoprotein, RNA (78-MER) | | Authors: | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
7NT6
 
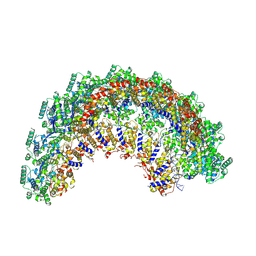 | | CryoEM structure of the Nipah virus nucleocapsid spiral clam-shaped assembly | | Descriptor: | Nucleoprotein, RNA (42-MER), RNA (48-MER) | | Authors: | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
