5CDN
 
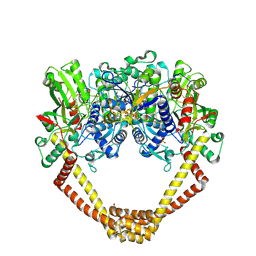 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
1WAM
 
 | |
5MG2
 
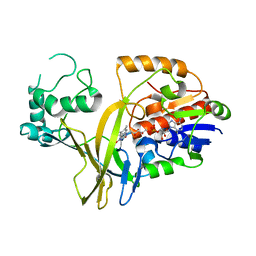
 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
2BI8
 
 | |
2BI7
 
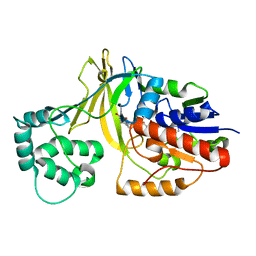 | | udp-galactopyranose mutase from Klebsiella pneumoniae oxidised FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Srikannathasan, V, Naismith, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mycobacteria Tuberculosis and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the Oxidised State and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the (Active) Reduced State.
J.Mol.Biol., 348, 2005
|
|
5BPV
 
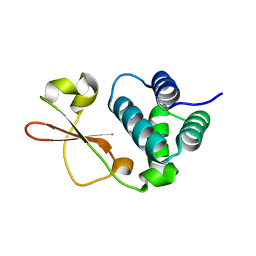 | | Crystal Structure of Zaire ebolavirus VP35 RNA binding domain mutant I278A | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Fadda, V, Cannas, V, Zinzula, L, Distinto, S, Daino, G.L, Bianco, G, Corona, A, Esposito, F, Alcaro, S, Maccioni, E, Tramontano, E, Taylor, G.L. | | Deposit date: | 2015-05-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure of Zaire ebolavirus VP35 RNA binding domain mutant I278A
to be published
|
|
5BQK
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL DOMAIN OF ICP27 PROTEIN FROM HSV-1 | | Descriptor: | ICP27, ZINC ION | | Authors: | Patel, V, Rajakannan, V, Dahlroth, S, Nordlund, P. | | Deposit date: | 2015-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-Terminal Domain of the Multifunctional ICP27 Protein from Herpes Simplex Virus 1.
J.Virol., 89, 2015
|
|
5CDM
 
 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDR
 
 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDP
 
 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5NF7
 
 | | Structure of Galectin-3 CRD in complex with compound 1 | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-N-acetyl-2-(acetylamino)-2-deoxy-beta-D-glucopyranosylamine | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Pilard, F.D, Teletchea, S, Ciesielski, F, Hannah, V.V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5LF9
 
 | | Crystal structure of human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22 | | Authors: | Tallant, C, Siejka, P, Mathea, S, Shrestha, L, Krojer, T, Srikannathasan, V, Elkins, J.M, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human NUDT22
To Be Published
|
|
4CO9
 
 | | Crystal structure of kynurenine formamidase from Bacillus anthracis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, KYNURENINE FORMAMIDASE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-27 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
4COB
 
 | | Crystal structure kynurenine formamidase from Pseudomonas aeruginosa | | Descriptor: | KYNURENINE FORMAMIDASE, ZINC ION | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
4AX2
 
 | | New Type VI-secreted toxins and self-resistance proteins in Serratia marcescens | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, RAP1B | | Authors: | English, G, Trunk, K, Rao, V.A, Srikannathasan, V, Fritsch, M.J, Guo, M, Hunter, W.N, Coulthurst, S.J. | | Deposit date: | 2012-06-07 | | Release date: | 2012-09-19 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New Secreted Toxins and Immunity Proteins Encoded within the Type Vi Secretion System Gene Cluster of Serratia Marcescens
Mol.Microbiol., 86, 2012
|
|
4CZ1
 
 | | Crystal structure of kynurenine formamidase from Bacillus anthracis complexed with 2-aminoacetophenone. | | Descriptor: | 2-aminoacetophenone, KYNURENINE FORMAMIDASE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-04-16 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
4COG
 
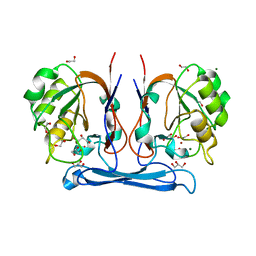 | | Crystal structure of kynurenine formamidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
1VL9
 
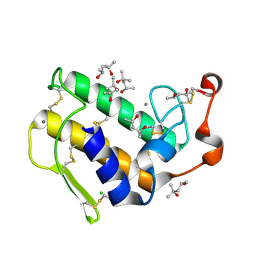 | | Atomic resolution (0.97A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Gayathri, D, Poi, M.-J, Tsai, M.-D, Dauter, M, Dauter, Z. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5NFC
 
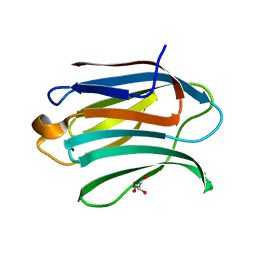 | | Structure of Galectin-3 CRD in complex with glycerol | | Descriptor: | GLYCEROL, Galectin-3 | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5NFB
 
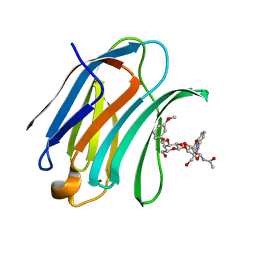 | | Structure of Galectin-3 CRD in complex with compound 4 | | Descriptor: | Galectin-3, ~{N}-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-acetamido-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-4-[(3-methoxyphenyl)methoxy]-3,5-bis(oxidanyl)oxan-2-yl]oxy-4-oxidanyl-oxan-3-yl]-3-methoxy-benzamide | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5NF9
 
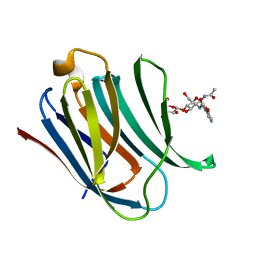 | | Structure of Galectin-3 CRD in complex with compound 2 | | Descriptor: | Galectin-3, ~{N}-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-acetamido-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4-oxidanyl-oxan-3-yl]-3-methoxy-benzamide | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
1VKQ
 
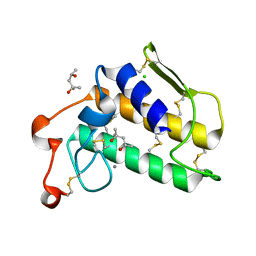 | | A re-determination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6A resolution using sulphur-SAS at 1.54A wavelength | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Yamane, T, Dauter, M, Dauter, Z. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redetermination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6 A resolution using sulfur-SAS at 1.54 A wavelength.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5NFA
 
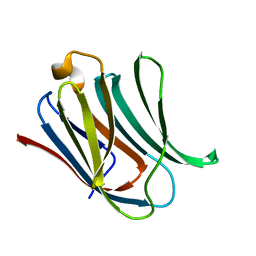 | | Structure of Galectin-3 CRD in complex with compound 3 | | Descriptor: | 3-O-[(3-methoxyphenyl)methyl]-beta-D-galactopyranose-(1-4)-N-acetyl-2-(acetylamino)-2-deoxy-beta-D-glucopyranosylamine, Galectin-3 | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5A3P
 
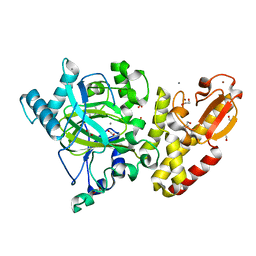 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FYB
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC1648 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Rotili, D, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-05 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc1648
To be Published
|
|
