7V9Z
 
 | |
7VA3
 
 | |
6VZO
 
 | |
6VZM
 
 | |
6VZN
 
 | |
6VZL
 
 | |
7SJR
 
 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
7VA6
 
 | |
7VA2
 
 | |
4NS5
 
 | | Crystal structure of human BS69 Bromo-Zinc finger-PWWP | | Descriptor: | ZINC ION, Zinc finger MYND domain-containing protein 11 | | Authors: | Wang, J.C, Qin, S, Li, F.D, Li, S, Zhang, W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human BS69 Bromo-ZnF-PWWP reveals its role in H3K36me3 nucleosome binding.
Cell Res., 24, 2014
|
|
8WA2
 
 | | cryo-EM structure of native mastigonemes isolated from Chlamydomonas reinhardtii at 3.0 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, J, Tao, H, Chen, J, Pan, J, Yan, C, Yan, N. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-guided discovery of protein and glycan components in native mastigonemes.
Cell, 187, 2024
|
|
3QAX
 
 | |
8T1R
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
5IFG
 
 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
4EQG
 
 | |
8FHN
 
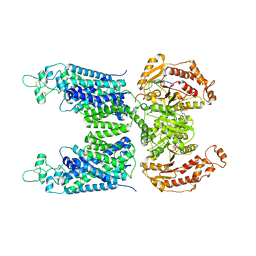 | | Cryo-EM structure of human NCC (class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Polythiazide, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHR
 
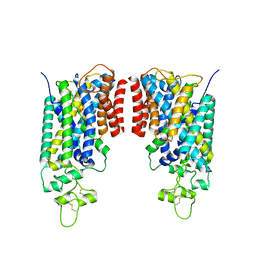 | | Cryo-EM structure of human NCC (class 3-3) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
4X23
 
 | |
8FHO
 
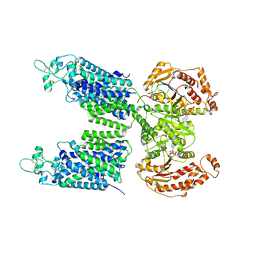 | | Cryo-EM structure of human NCC (class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Polythiazide, SODIUM ION, ... | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHP
 
 | | Cryo-EM structure of human NCC (class 3-1) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHT
 
 | | Cryo-EM structure of human NCC | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 12 member 3 | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHQ
 
 | | Cryo-EM structure of human NCC (class 3-2) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
5WEU
 
 | | Crystal Structure of H2-Dd with disulfide-linked 10mer peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Envelope glycoprotein gp160, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
