4C51
 
 | | Crystal Structure of the Catalase-Peroxidase (KatG) R418L mutant from Mycobacterium Tuberculosis | | Descriptor: | CATALASE-PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, alpha-D-glucopyranose | | Authors: | Hersleth, H.-P, Zhao, X, Magliozzo, R.S, Andersson, K.K. | | Deposit date: | 2013-09-10 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Access Channel Residues Ser315 and Asp137 in Mycobacterium Tuberculosis Catalase-Peroxidase (Katg) Control Peroxidatic Activation of the Pro-Drug Isoniazid.
Chem.Commun.(Camb.), 49, 2013
|
|
5VQW
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acrylamide (JLJ685), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)prop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Kudalkar, S.N, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQX
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQS
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acrylamide (JLJ685), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)prop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQU
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide (JLJ683), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Petrova, Z.O, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQR
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQZ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Buckingham, A.B, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQT
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | 2-chloro-N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Czyzyk, D.J, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQV
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQQ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide (JLJ683), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQY
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TER
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Conformation, and Crystallography of 2-Naphthyl Phenyl Ethers as Potent Anti-HIV Agents.
ACS Med Chem Lett, 7, 2016
|
|
3DG8
 
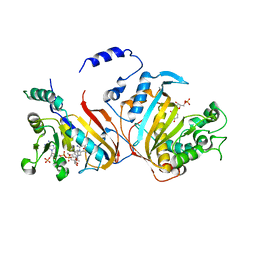 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with RJF670, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, N-(3,5-dimethoxyphenyl)imidodicarbonimidic diamide, ... | | Authors: | Dasgupta, T, Chitnumsub, P, Maneeruttanarungroj, C, Kamchonwongpaisan, S, Nichols, S, Lyons, T.M, Tirado-Rives, J, Jorgensen, W.L, Yuthavong, Y, Anderson, K.S. | | Deposit date: | 2008-06-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Exploiting structural analysis, in silico screening, and serendipity to identify novel inhibitors of drug-resistant falciparum malaria.
Acs Chem.Biol., 4, 2009
|
|
3DL5
 
 | | Crystal Structure of the A287F Active Site Mutant of TS-DHFR from Cryptosporidium hominis | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DIHYDROFOLIC ACID, ... | | Authors: | Vargo, M.A, Martucci, W.E, Anderson, K.S. | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase.
Biochemistry, 47, 2008
|
|
1BXN
 
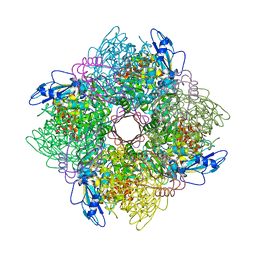 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
1H0O
 
 | |
1H0N
 
 | |
3DL6
 
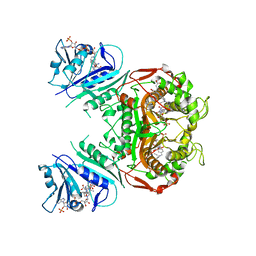 | | Crystal Structure of the A287F/S290G Active Site Mutant of TS-DHFR from Cryptosporidium hominis | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DIHYDROFOLIC ACID, ... | | Authors: | Martucci, W.E, Vargo, M.A, Anderson, K.S. | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase.
Biochemistry, 47, 2008
|
|
3DGA
 
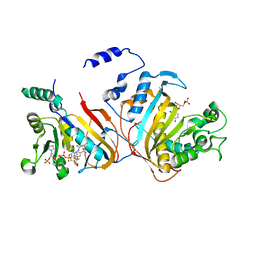 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with RJF01302, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, N-[2-chloro-5-(trifluoromethyl)phenyl]imidodicarbonimidic diamide, ... | | Authors: | Dasgupta, T, Chitnumsub, P, Maneeruttanarungroj, C, Kamchonwongpaisan, S, Nichols, S, Lyons, T.M, Tirado-Rives, J, Jorgensen, W.L, Yuthavong, Y, Anderson, K.S. | | Deposit date: | 2008-06-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploiting structural analysis, in silico screening, and serendipity to identify novel inhibitors of drug-resistant falciparum malaria.
Acs Chem.Biol., 4, 2009
|
|
4J0N
 
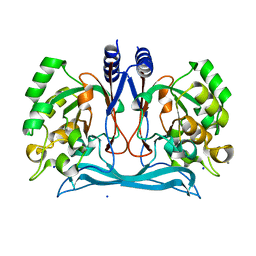 | | Crystal structure of a manganese dependent isatin hydrolase | | Descriptor: | CALCIUM ION, Isatin hydrolase B, MANGANESE (II) ION, ... | | Authors: | Bjerregaard-Andersen, K, Sommer, T, Jensen, J.K, Jochimsen, B, Etzerodt, M, Morth, J.P. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A proton wire and water channel revealed in the crystal structure of isatin hydrolase.
J.Biol.Chem., 289, 2014
|
|
1ACA
 
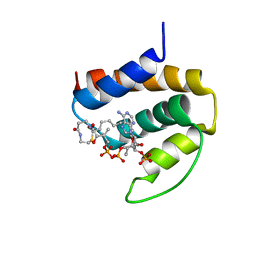 | | THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX BETWEEN ACYL-COENZYME A BINDING PROTEIN AND PALMITOYL-COENZYME A | | Descriptor: | ACYL-COENZYME A BINDING PROTEIN, COENZYME A, PALMITIC ACID | | Authors: | Kragelund, B.B, Andersen, K.V, Madsen, J.C, Knudsen, J, Poulsen, F.M. | | Deposit date: | 1992-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the complex between acyl-coenzyme A binding protein and palmitoyl-coenzyme A.
J.Mol.Biol., 230, 1993
|
|
3R9U
 
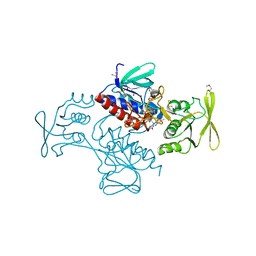 | | Thioredoxin-disulfide reductase from Campylobacter jejuni. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, K.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Thioredoxin-disulfide reductase from Campylobacter jejuni.
To be Published
|
|
3PRX
 
 | | Structure of Complement C5 in Complex with CVF and SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | Authors: | Laursen, N.S, Andersen, G.R, Sottrup-Jensen, L, Andersen, K.R, Spillner, E, Braren, I. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Embo J., 30, 2011
|
|
4A3H
 
 | | 2',4' DINITROPHENYL-2-DEOXY-2-FLURO-B-D-CELLOBIOSIDE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS AT 1.6 A RESOLUTION | | Descriptor: | 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-BETA-D-CELLOBIOSIDE, PROTEIN (ENDOGLUCANASE) | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
1A3H
 
 | |
