5TK4
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein from Bacillus anthracis | | Descriptor: | Cytochrome B | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein from
Bacillus anthracis
To Be Published
|
|
5TLS
 
 | | 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD | | Descriptor: | (2S)-4-(6-amino-9H-purin-9-yl)-2-hydroxybutanoic acid, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD
To Be Published
|
|
3OJC
 
 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
5TKW
 
 | | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae. | | Descriptor: | FORMIC ACID, Type II secretion system protein L | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae.
To Be Published
|
|
5TF3
 
 | | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, Putative membrane protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Anderson, W.F, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis
To Be Published
|
|
3Q4G
 
 | | Structure of NAD synthetase from Vibrio cholerae | | Descriptor: | CALCIUM ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of NAD synthetase from Vibrio cholerae
TO BE PUBLISHED
|
|
5TK2
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Cytochrome B, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from
Bacillus anthracis
To Be Published
|
|
3RUY
 
 | | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis | | Descriptor: | Ornithine aminotransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis
To be Published
|
|
2GX8
 
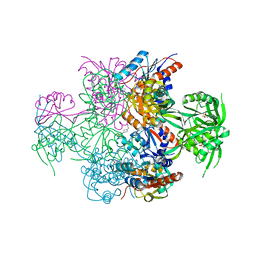 | | The Crystal Structure of Bacillus cereus protein related to NIF3 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NIF3-related protein, ... | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-08 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution crystal structure of Bacillus cereus Nif3-family protein YqfO reveals a conserved dimetal-binding motif and a regulatory domain
Protein Sci., 16, 2007
|
|
2FOR
 
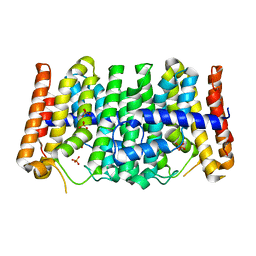 | | Crystal Structure of the Shigella flexneri Farnesyl Pyrophosphate Synthase Complex with an Isopentenyl Pyrophosphate | | Descriptor: | Geranyltranstransferase, ISOPENTYL PYROPHOSPHATE, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-13 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Shigella flexneri Farnesyl Pyrophosphate Synthase Complex with an Isopentenyl Pyrophosphate
To be Published
|
|
2FZV
 
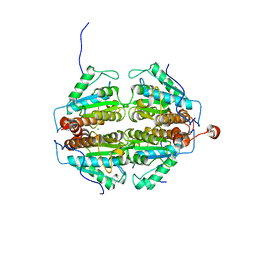 | | Crystal Structure of an apo form of a Flavin-binding Protein from Shigella flexneri | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative arsenical resistance protein | | Authors: | Vorontsov, I.I, Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-10 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity
Protein Sci., 16, 2007
|
|
2H1I
 
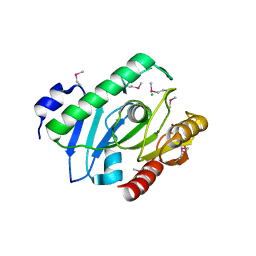 | | Crystal Structure of the Bacillus cereus Carboxylesterase | | Descriptor: | CALCIUM ION, CHLORIDE ION, Carboxylesterase, ... | | Authors: | Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-16 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Bacillus cereus Carboxylesterase
To be Published
|
|
2GJV
 
 | | Crystal Structure of a Protein of Unknown Function from Salmonella typhimurium | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative cytoplasmic protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-31 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of a Hypothetical Protein from Salmonella typhimurium
To be Published
|
|
2I0Z
 
 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
1XA0
 
 | | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus | | Descriptor: | CHLORIDE ION, Putative NADPH Dependent oxidoreductases, SULFATE ION | | Authors: | Brunzelle, J.S, Sommerhalter, M, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus
To be Published
|
|
1VR4
 
 | | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus | | Descriptor: | hypothetical protein APC22750 | | Authors: | Yang, X, Brunzelle, J.S, McNamara, L.K, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus
To be Published
|
|
4YPD
 
 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
4ZWL
 
 | | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-19 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289
To be Published
|
|
1VRK
 
 | |
1XPJ
 
 | | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae | | Descriptor: | L(+)-TARTARIC ACID, MERCURY (II) ION, hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC26283 from Vibrio cholerae
To be Published
|
|
4YO4
 
 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment phthalazine | | Descriptor: | ACETATE ION, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
1X7F
 
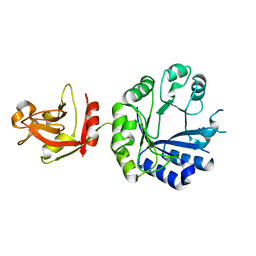 | | Crystal structure of an uncharacterized B. cereus protein | | Descriptor: | outer surface protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-13 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an uncharacterized B. cereus protein
To be Published
|
|
1Y2I
 
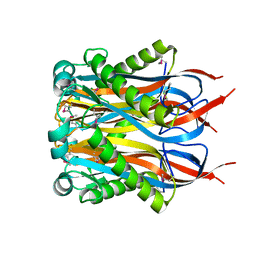 | | Crystal Structure of MCSG Target APC27401 from Shigella flexneri | | Descriptor: | Hypothetical Protein S0862 | | Authors: | Brunzelle, J.S, Minasov, G, Yang, X, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-22 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC27401 from Shigella flexneri
To be Published
|
|
1DUG
 
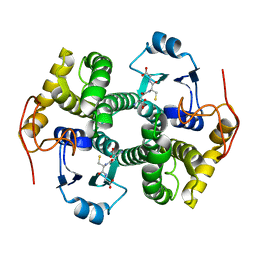 | | STRUCTURE OF THE FIBRINOGEN G CHAIN INTEGRIN BINDING AND FACTOR XIIIA CROSSLINKING SITES OBTAINED THROUGH CARRIER PROTEIN DRIVEN CRYSTALLIZATION | | Descriptor: | GLUTATHIONE, chimera of GLUTATHIONE S-TRANSFERASE-synthetic LINKEr-C-TERMINAL FIBRINOGEN GAMMA CHAIN | | Authors: | Ware, S, Donahue, J.P, Hawiger, J, Anderson, W.F. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the fibrinogen gamma-chain integrin binding and factor XIIIa cross-linking sites obtained through carrier protein driven crystallization.
Protein Sci., 8, 1999
|
|
6MUQ
 
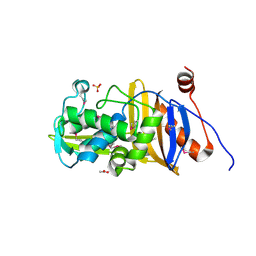 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | Descriptor: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
