3KBO
 
 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
4HS7
 
 | | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG. | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bacterial extracellular solute-binding protein, putative, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG.
TO BE PUBLISHED
|
|
3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4HTF
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase from Escherichia coli in complex with S-adenosylmethionine. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase from Escherichia coli in complex with S-adenosylmethionine.
To be Published
|
|
3KB8
 
 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
4HVN
 
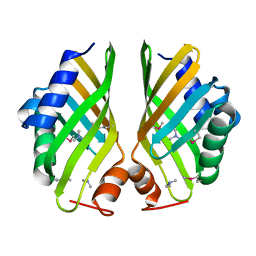 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 in complex with Trimethylamine. | | Descriptor: | N,N-dimethylmethanamine, hypothetical protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
4HUS
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with virginiamycin M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
4HN3
 
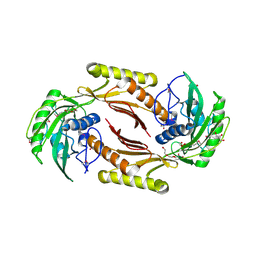 | | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e
To be Published
|
|
4I62
 
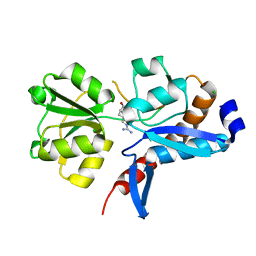 | | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Stogios, P.J, Kudritska, M, Wawrzak, Z, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine
To be Published
|
|
3KQF
 
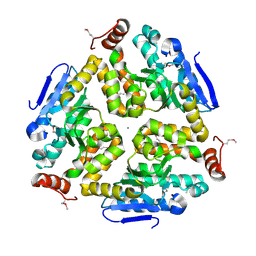 | | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Minasov, G, Halavaty, A, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-17 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis.
TO BE PUBLISHED
|
|
4HUR
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
3L07
 
 | | Methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bifunctional protein folD, ... | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-09 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis.
To be Published
|
|
3L2I
 
 | | 1.85 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2. | | Descriptor: | 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.
Biochemistry, 50, 2011
|
|
6U60
 
 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | Authors: | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
4HW8
 
 | | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose. | | Descriptor: | Bacterial extracellular solute-binding protein, putative, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose.
TO BE PUBLISHED
|
|
4IIW
 
 | | 2.6 Angstrom Crystal Structure of Putative yceG-like Protein lmo1499 from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Lmo1499 protein, SULFATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of Putative yceG-like Protein lmo1499 from Listeria monocytogenes
TO BE PUBLISHED
|
|
4IFA
 
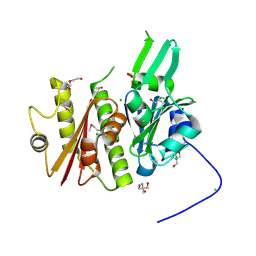 | | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Shatsman, S, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames
To be Published
|
|
3L93
 
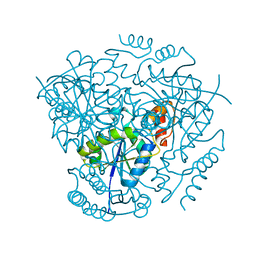 | | Phosphopantetheine adenylyltransferase from Yersinia pestis. | | Descriptor: | FORMIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Osipiuk, J, Maltseva, N, Makowska-grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | X-ray crystal structure of phosphopantetheine adenylyltransferase from Yersinia pestis.
To be Published
|
|
3LHQ
 
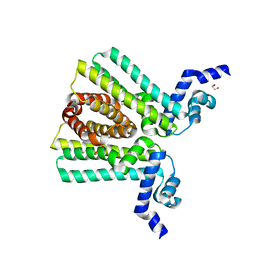 | | DNA-binding transcriptional repressor AcrR from Salmonella typhimurium. | | Descriptor: | 1,2-ETHANEDIOL, AcrAB operon repressor (TetR/AcrR family), DI(HYDROXYETHYL)ETHER | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of DNA-binding transcriptional repressor AcrR from Salmonella typhimurium.
To be Published
|
|
3LAY
 
 | | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Zinc resistance-associated protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
4IIN
 
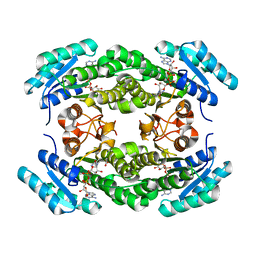 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+ | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+
To be Published
|
|
4IJK
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), SODIUM ION | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I.G, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695
To be Published
|
|
3LER
 
 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-15 | | Release date: | 2010-01-26 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4IIU
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution
To be Published
|
|
4ICH
 
 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
