4WS5
 
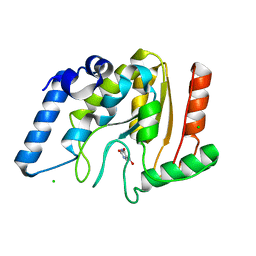 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-nitrouracil, Form III | | Descriptor: | 5-nitrouracil, CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRU
 
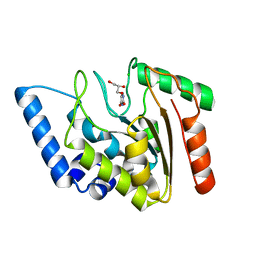 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form II | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRW
 
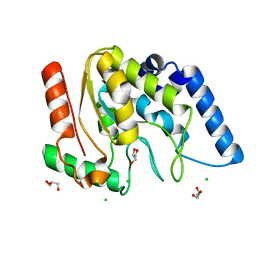 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form IV | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRY
 
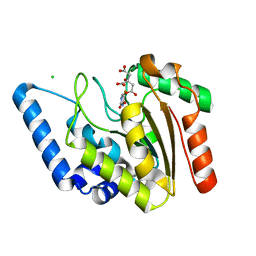 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil(B), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WPK
 
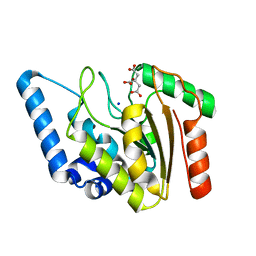 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form I | | Descriptor: | CITRIC ACID, SODIUM ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5NUI
 
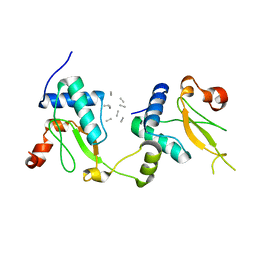 | | Crystal structure of SIVmac239 Nef in an ExxxLM endocytic sorting motif bound state | | Descriptor: | Protein Nef, SER-GLN-ILE-LYS-ARG-LEU-LEU-SER | | Authors: | Manrique, S, Horenkamp, F.A, Anand, K, Geyer, M. | | Deposit date: | 2017-04-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3.
Nat Commun, 8, 2017
|
|
6Z2G
 
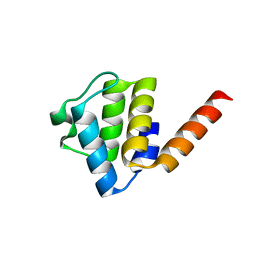 | | Crystal structure of human NLRP9 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Marleaux, M, Anand, K, Geyer, M. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly.
Febs Lett., 594, 2020
|
|
5NUH
 
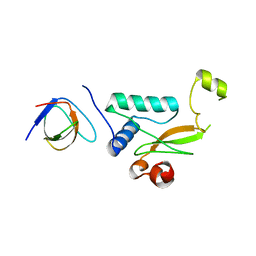 | |
4NST
 
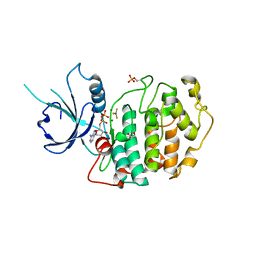 | | Crystal structure of human Cdk12/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Boesken, C.A, Farnung, L, Anand, K, Geyer, M. | | Deposit date: | 2013-11-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and substrate specificity of human Cdk12/Cyclin K.
Nat Commun, 5, 2014
|
|
4YDH
 
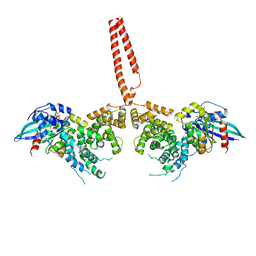 | | The structure of human FMNL1 N-terminal domains bound to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Formin-like protein 1, MAGNESIUM ION, ... | | Authors: | Kuhn, S, Anand, K, Geyer, M. | | Deposit date: | 2015-02-22 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation.
Nat Commun, 6, 2015
|
|
7O7K
 
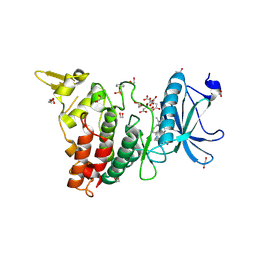 | | Crystal structure of the human DYRK1A kinase domain bound to abemaciclib | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
5EFQ
 
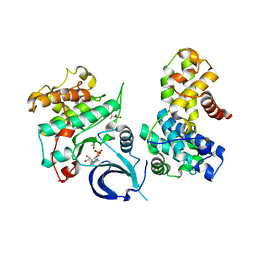 | | Crystal structure of human Cdk13/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin-K, ... | | Authors: | Hoenig, D, Greifenberg, A.K, Anand, K, Geyer, M. | | Deposit date: | 2015-10-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of the Cdk13/Cyclin K Complex.
Cell Rep, 14, 2016
|
|
2HKO
 
 | | Crystal structure of LSD1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1 | | Authors: | Chen, Y, Yang, Y.T, Wang, F, Yanane, K, Zhang, Y, Lei, M. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human histone lysine-specific demethylase 1 (LSD1).
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4B2Z
 
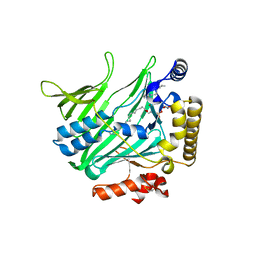 | | Structure of Osh6 in complex with phosphatidylserine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Maeda, K, Anand, K, Chiapparino, A, Kumar, A, Poletto, M, Kaksonen, M, Gavin, A.C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins
Nature, 501, 2013
|
|
7N9U
 
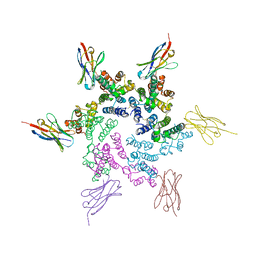 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7N9X
 
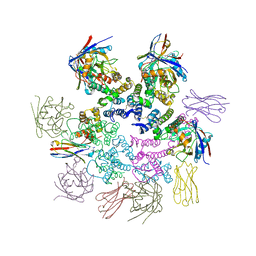 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.511 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7O7I
 
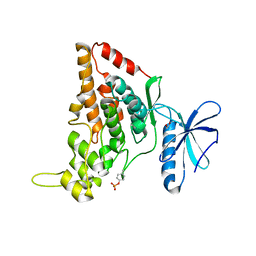 | |
7O7J
 
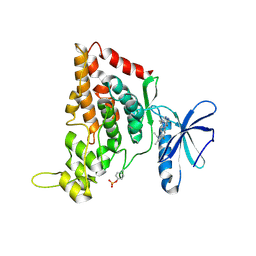 | | Crystal structure of the human HIPK3 kinase domain bound to abemaciclib | | Descriptor: | Homeodomain-interacting protein kinase 3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
1UK4
 
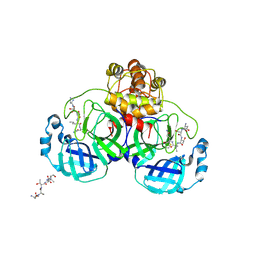 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1TC8
 
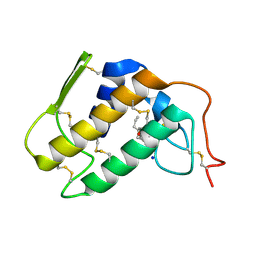 | | Crystal structure of Krait-venom phospholipase A2 in a complex with a natural fatty acid tridecanoic acid | | Descriptor: | N-TRIDECANOIC ACID, SODIUM ION, phospholipase A2 isoform 1 | | Authors: | Singh, G, Jasti, J, Saravanan, K, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex formed between a group I phospholipase A2 and a naturally occurring fatty acid at 2.7 A resolution
PROTEIN SCI., 14, 2005
|
|
1TFV
 
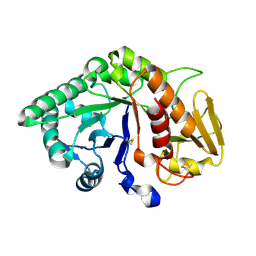 | | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40) SECRETED DURING INVOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, mammary gland protein 40 | | Authors: | Bilgrami, S, Saravanan, K, Yadav, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40)
SECRETED DURING INVOLUTION
To be Published
|
|
1UJ1
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK2
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 | | Descriptor: | 3C-LIKE PROTEINASE | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK3
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1LJY
 
 | | Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MGP-40 | | Authors: | Mohanty, A.K, Singh, G, Paramasivam, M, Saravanan, K, Jabeen, T, Sharma, S, Yadav, S, Kaur, P, Kumar, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-04-23 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Novel Regulatory 40 kDa Mammary Gland Protein (MGP-40) secreted during Involution
J.Biol.Chem., 278, 2003
|
|
