7VE3
 
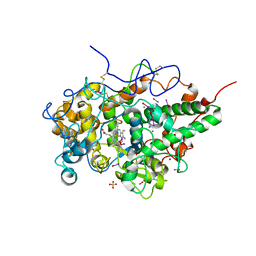 | | Structure of the complex of sheep lactoperoxidase with hypoiodite at 2.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, P.K, Yamini, S, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence of the oxidation of iodide ion into hyper-reactive hypoiodite ion by mammalian heme lactoperoxidase.
Protein Sci., 31, 2022
|
|
3LUA
 
 | |
3LTO
 
 | |
2I3O
 
 | |
3LL3
 
 | |
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
3NHM
 
 | |
3NPK
 
 | |
3NYW
 
 | |
2IMR
 
 | |
3MAB
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM LISTERIA MONOCYTOGENES, Triclinic FORM | | Descriptor: | uncharacterized protein | | Authors: | Madegowda, M, Chruszcz, M, Minor, W, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of an uncharacterized protein from listeria monocytogenes
To be Published
|
|
3MIZ
 
 | |
3MPO
 
 | |
3MSR
 
 | |
3LKB
 
 | |
3MSY
 
 | |
2I6E
 
 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
3M1Y
 
 | |
2I76
 
 | | Crystal structure of protein TM1727 from Thermotoga maritima | | Descriptor: | Hypothetical protein, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Madegowda, M, Eswaramoorthy, S, Seetharaman, J, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of hypothetical protein TM1727 from Thermatoga maritima
TO BE PUBLISHED
|
|
3NAS
 
 | |
3NQB
 
 | |
3MGG
 
 | |
3MKC
 
 | |
3M2T
 
 | |
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
