6USF
 
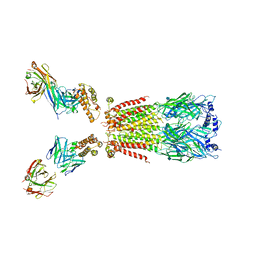 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
6UR8
 
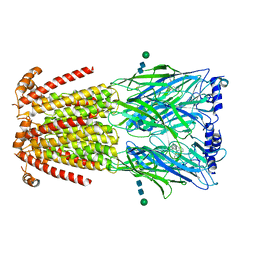 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera of soluble cytochrome b562 (BRIL) and neuronal acetylcholine receptor subunit alpha-4, Neuronal acetylcholine receptor subunit beta-2, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCR
 
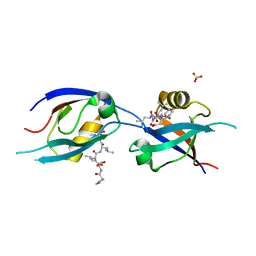 | | MLLT4/Afadin PDZ domain in complex with the C-terminal peptide from protein E of SARS-CoV-2 | | Descriptor: | Afadin, Envelope small membrane protein, SULFATE ION | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
3JA6
 
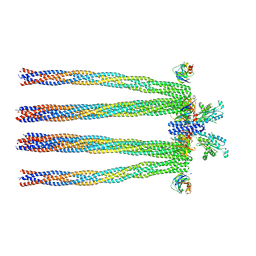 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
7QCS
 
 | | PALS1/MPP5 PDZ domain in complex with SARS-CoV-2_E PBM peptide | | Descriptor: | Envelope small membrane protein, Protein PALS1 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
5UOT
 
 | |
