2QMP
 
 | | Crystal Structure of HIV-1 protease complexed with PL-100 | | Descriptor: | N-[(5S)-5-{[(4-aminophenyl)sulfonyl](isobutyl)amino}-6-hydroxyhexyl]-Nalpha-(methoxycarbonyl)-beta-phenyl-L-phenylalaninamide, Pol polyprotein | | Authors: | Allison, T.J. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of HIV-1 protease complexed with PL-100
To be Published
|
|
1A62
 
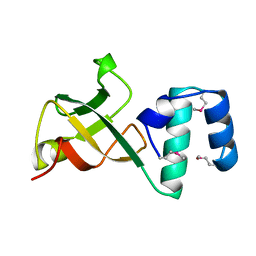 | | CRYSTAL STRUCTURE OF THE RNA-BINDING DOMAIN OF THE TRANSCRIPTIONAL TERMINATOR PROTEIN RHO | | Descriptor: | RHO | | Authors: | Allison, T.J, Wood, T.C, Briercheck, D.M, Rastinejad, F, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the RNA-binding domain from transcription termination factor rho.
Nat.Struct.Biol., 5, 1998
|
|
3H0B
 
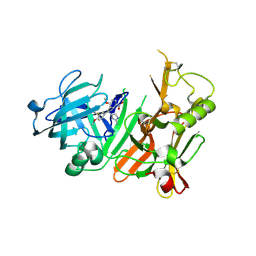 | | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class | | Descriptor: | 4-[(1S)-1-(3-fluoro-4-methoxyphenyl)-2-(2-methoxy-5-nitrophenyl)ethyl]-1H-imidazol-2-amine, Beta-secretase 1 | | Authors: | Allison, T.J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class: pH dependence on binding activity part 1.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EXO
 
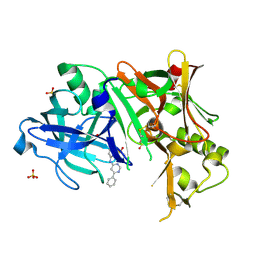 | | Crystal structure of BACE1 bound to inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{2-methyl-5-[(6-phenylpyrimidin-4-yl)amino]phenyl}methanesulfonamide, ... | | Authors: | Allison, T.J. | | Deposit date: | 2008-10-16 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a small molecule beta-secretase inhibitor that binds without catalytic aspartate engagement.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1HXM
 
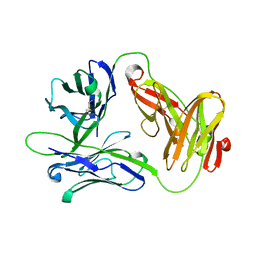 | | Crystal Structure of a Human Vgamma9/Vdelta2 T Cell Receptor | | Descriptor: | GAMMA-DELTA T-CELL RECEPTOR, SULFATE ION | | Authors: | Allison, T.J, Winter, C.C, Fournie, J.J, Bonneville, M, Garboczi, D.N. | | Deposit date: | 2001-01-16 | | Release date: | 2001-06-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of a human gammadelta T-cell antigen receptor.
Nature, 411, 2001
|
|
2ZJM
 
 | |
1HNA
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1A63
 
 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | Descriptor: | RHO | | Authors: | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
1HNB
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HNC
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1YPY
 
 | | Crystal Structure of Vaccinia Virus L1 protein | | Descriptor: | Virion membrane protein | | Authors: | Su, H.P, Garman, S.C, Allison, T.J, Fogg, C, Moss, B, Garboczi, D.N. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The 1.51-Angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
