6UPC
 
 | |
1XYC
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYB
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
3E84
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
1XYA
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
6XCB
 
 | |
6XCC
 
 | |
6XCD
 
 | |
2ILP
 
 | |
2IMB
 
 | |
2IMC
 
 | |
2IMA
 
 | |
2FUE
 
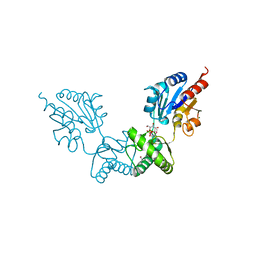 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2FUC
 
 | | Human alpha-Phosphomannomutase 1 with Mg2+ cofactor bound | | Descriptor: | MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
6XCE
 
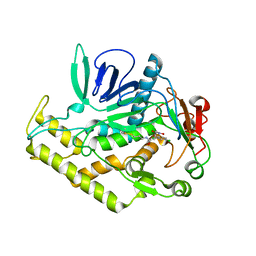 | |
6XCF
 
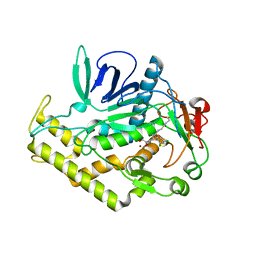 | |
2YBD
 
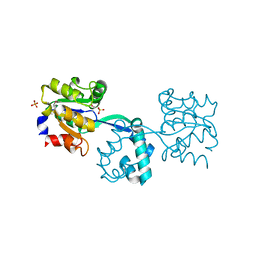 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
1CQD
 
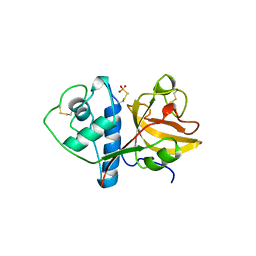 | | THE 2.1 ANGSTROM STRUCTURE OF A CYSTEINE PROTEASE WITH PROLINE SPECIFICITY FROM GINGER RHIZOME, ZINGIBER OFFICINALE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PROTEASE II), ... | | Authors: | Choi, K.H, Laursen, R.A, Allen, K.N. | | Deposit date: | 1999-06-15 | | Release date: | 1999-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A structure of a cysteine protease with proline specificity from ginger rhizome, Zingiber officinale.
Biochemistry, 38, 1999
|
|
3BH2
 
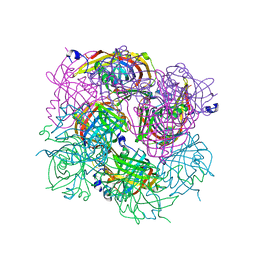 | |
3BGT
 
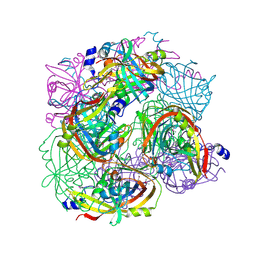 | | Structural Studies of Acetoacetate Decarboxylase | | Descriptor: | Probable acetoacetate decarboxylase | | Authors: | Ho, M, Allen, K.N. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The origin of the electrostatic perturbation in acetoacetate decarboxylase.
Nature, 459, 2009
|
|
3BH3
 
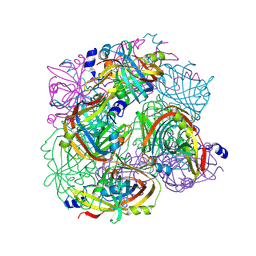 | |
3BOK
 
 | |
3BV4
 
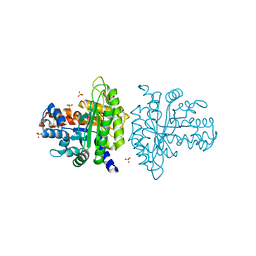 | | Crystal structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase A, SULFATE ION | | Authors: | Sherawat, M, Tolan, D.R, Allen, K.N. | | Deposit date: | 2008-01-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3BON
 
 | |
3BOO
 
 | |
