6MPO
 
 | |
6MNT
 
 | |
6D53
 
 | |
6D5Z
 
 | |
1Z3I
 
 | | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | | Descriptor: | SULFATE ION, ZINC ION, similar to RAD54-like | | Authors: | Thoma, N.H, Czyzewski, B.K, Alexeev, A.A, Mazin, A.V, Kowalczykowski, S.C, Pavletich, N.P. | | Deposit date: | 2005-03-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1IK1
 
 | | Solution Structure of an RNA Hairpin from HRV-14 | | Descriptor: | 5'-R(*GP*GP*UP*AP*CP*UP*AP*UP*GP*UP*AP*CP*CP*A)-3' | | Authors: | Huang, H, Alexandrov, A, Chen, X, Barnes III, T.W, Zhang, H, Dutta, K, Pascal, S.M. | | Deposit date: | 2001-05-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin from HRV-14.
Biochemistry, 40, 2001
|
|
1PZ9
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1PZ7
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1Q56
 
 | | NMR structure of the B0 isoform of the agrin G3 domain in its Ca2+ bound state | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-08-06 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding
Structure, 12, 2004
|
|
1AQ5
 
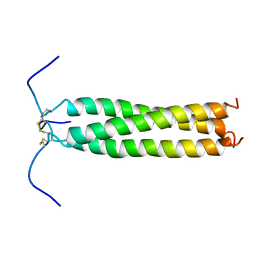 | | HIGH-RESOLUTION SOLUTION NMR STRUCTURE OF THE TRIMERIC COILED-COIL DOMAIN OF CHICKEN CARTILAGE MATRIX PROTEIN, 20 STRUCTURES | | Descriptor: | CARTILAGE MATRIX PROTEIN | | Authors: | Dames, S.A, Wiltscheck, R, Kammerer, R.A, Engel, J, Alexandrescu, A.T. | | Deposit date: | 1997-08-07 | | Release date: | 1998-02-11 | | Last modified: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a parallel homotrimeric coiled coil.
Nat.Struct.Biol., 5, 1998
|
|
1QOJ
 
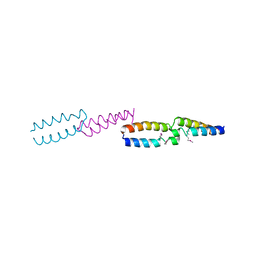 | | Crystal Structure of E.coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. | | Descriptor: | UVRB | | Authors: | Sohi, M, Alexandrovich, A, Moolenaar, G, Visse, R, Goosen, N, Vernede, X, Fontecilla-Camps, J, Champness, J, Sanderson, M.R. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-10 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of E.Coli Uvrb C-Terminal Domain, and a Model for Uvrb-Uvrc Interaction
FEBS Lett., 465, 2000
|
|
1PZ8
 
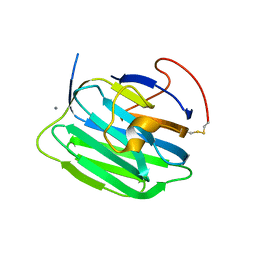 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
2N67
 
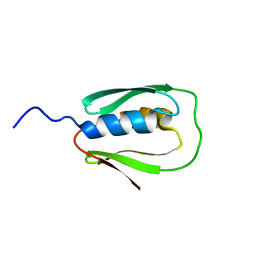 | |
2M5S
 
 | | High-resolution NMR structure and cryo-EM imaging support multiple functional roles for the accessory I-domain of phage P22 coat protein | | Descriptor: | Coat protein | | Authors: | Rizzo, A.A, Suhanovsky, M.M, Baker, M.L, Fraser, L.C.R, Jones, L.M, Rempel, D.L, Gross, M.L, Chiu, W, Alexandrescu, A.T, Teschke, C.M. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling.
Structure, 22, 2014
|
|
2OVN
 
 | |
1TM6
 
 | |
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
2LPM
 
 | |
2KB8
 
 | |
2M98
 
 | |
4LH8
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
