7RR9
 
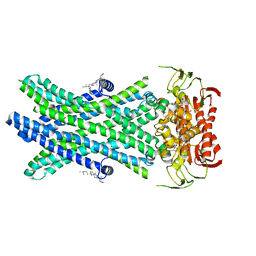 | | Cryo-EM Structure of Nanodisc reconstituted ABCD1 in nucleotide bound outward open conformation | | Descriptor: | ATP-binding cassette sub-family D member 1, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human peroxisomal fatty acid transporter ABCD1 in a lipid environment
Commun Biol, 5, 2022
|
|
7RRA
 
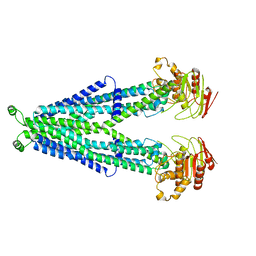 | |
8EEB
 
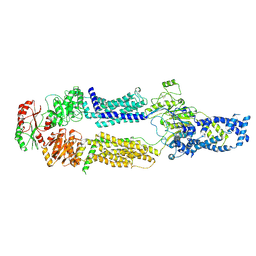 | | Cryo-EM structure of human ABCA7 in Digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EOP
 
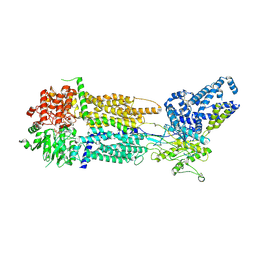 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EE6
 
 | | Cryo-EM Structure of human ABCA7 in PE/Ch nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EDW
 
 | | Cryo-EM Structure of human ABCA7 in BPL/Ch Nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
6QEE
 
 | |
6QEX
 
 | |
6FN1
 
 | | Zosuquidar and UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Human-mouse chimeric ABCB1 (ABCBHM), UIC2 Antigen Binding Fragment Heavy Chain, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FN4
 
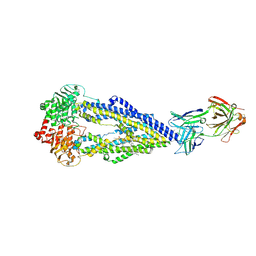 | | Apo form of UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Apo form of Human-mouse chimeric ABCB1 (ABCB1HM) in complex with Antigen binding fragment of UIC2., ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ZRP
 
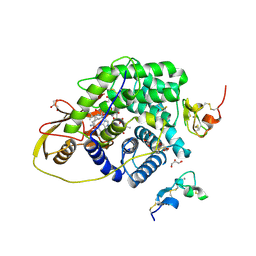 | | TC:CD320 | | Descriptor: | CALCIUM ION, CD320 antigen, CYANOCOBALAMIN, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2015-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of transcobalamin recognition by human CD320 receptor.
Nat Commun, 7, 2016
|
|
4ZRQ
 
 | |
2Q69
 
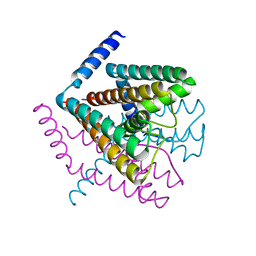 | | Crystal Structure of Nak channel D66N mutant | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Alam, A, Shi, N, Jiang, Y. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into Ca2+ specificity in tetrameric cation channels.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q6A
 
 | | Crystal Structure of Nak channel D66E mutant | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Alam, A, Shi, N, Jiang, Y. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into Ca2+ specificity in tetrameric cation channels.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q67
 
 | | Crystal Structure of Nak channel D66A mutant | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Alam, A, Shi, N, Jiang, Y. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into Ca2+ specificity in tetrameric cation channels.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q68
 
 | | Crystal Structure of Nak channel D66A, S70E double mutants | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Alam, A, Shi, N, Jiang, Y. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into Ca2+ specificity in tetrameric cation channels.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
9CTG
 
 | | SapNP Reconstituted Human ABCB1 bound to ATP gammaS | | Descriptor: | ATP-dependent translocase ABCB1, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kurre, D, Alam, A. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into binding-site access and ligand recognition by human ABCB1.
Embo J., 44, 2025
|
|
9CR8
 
 | | SapNP reconstituted human ABCB1 | | Descriptor: | ATP-dependent translocase ABCB1, UNKNOWN BRANCHED FRAGMENT OF PHOSPHOLIPID | | Authors: | Kurre, D, Alam, A. | | Deposit date: | 2024-07-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into binding-site access and ligand recognition by human ABCB1.
Embo J., 44, 2025
|
|
9CTC
 
 | |
9CTF
 
 | |
6S7P
 
 | | Nucleotide bound ABCB4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Olsen, J.A, Alam, A, Kowal, J, Stieger, B, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human lipid exporter ABCB4 in a lipid environment.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3E86
 
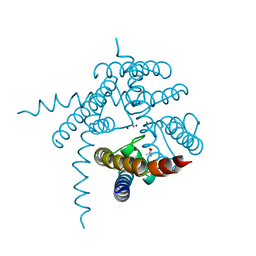 | |
3E83
 
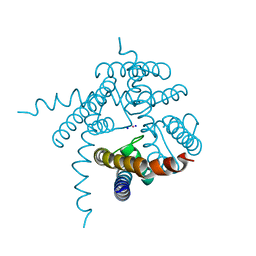 | | Crystal Structure of the the open NaK channel pore | | Descriptor: | CESIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Jiang, Y, Alam, A. | | Deposit date: | 2008-08-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of ion selectivity in the NaK channel
Nat.Struct.Mol.Biol., 16, 2009
|
|
3E8F
 
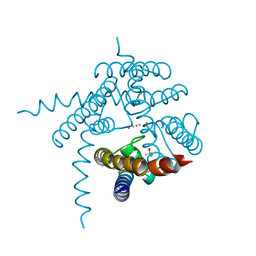 | |
3E8B
 
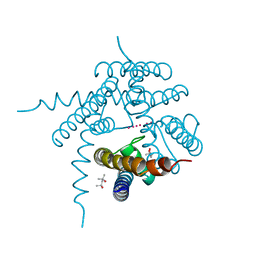 | |
