3GFH
 
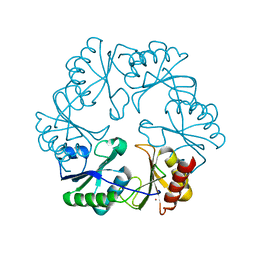 | |
5GJE
 
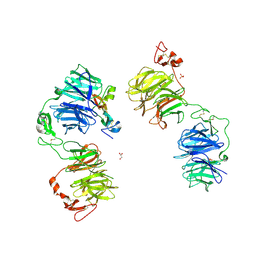 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
1RCH
 
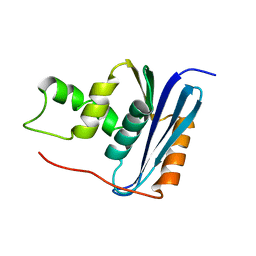 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
1RRB
 
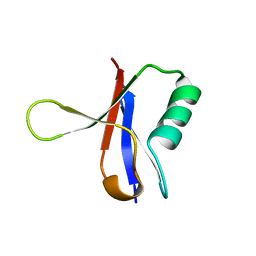 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
1AYG
 
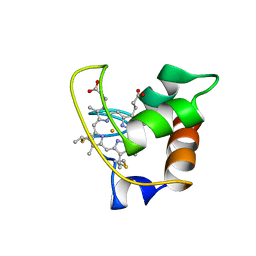 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1C9F
 
 | |
1MH5
 
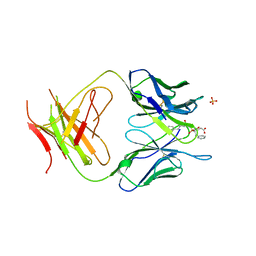 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJ8
 
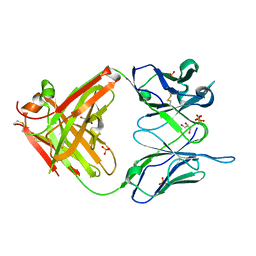 | | High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-126, PHOSPHATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
6AB6
 
 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AB5
 
 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
1UKJ
 
 | | Detailed structure of L-Methionine-Lyase from Pseudomonas putida | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2003-08-24 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detailed structure of L-Methionine -Lyase from Pseudomonas putida
To be Published
|
|
1MJJ
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG | | Descriptor: | IMMUNOGLOBULIN MS6-12, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1A05
 
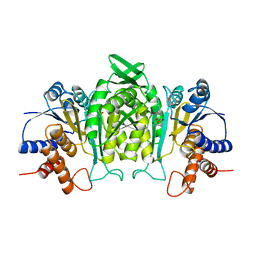 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
5XOF
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide I | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Nitric oxide synthase, ... | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, H, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
1MJU
 
 | | 1.22 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-12 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
7VR9
 
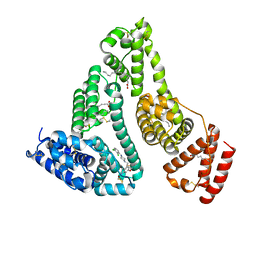 | | Crystal structure of human serum albumin complex with aripiprazole and myristic acid | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, MYRISTIC ACID, Serum albumin | | Authors: | Kawai, A, Yamasaki, K, Otagiri, M. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of Myristate on the Induced Circular Dichroism Spectra of Aripiprazole Bound to Human Serum Albumin: A Structural-Chemical Investigation
Acs Omega, 7, 2022
|
|
1F2H
 
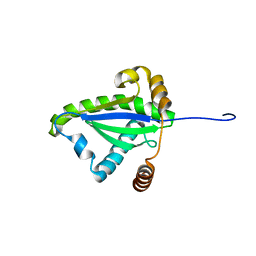 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE TNFR1 ASSOCIATED PROTEIN, TRADD. | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN | | Authors: | Tsao, D, McDonaugh, T, Malakian, K, Xu, G.-Y, Telliez, J.-B, Hsu, H, Lin, L.-L. | | Deposit date: | 2000-05-24 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway.
Mol.Cell, 5, 2000
|
|
1MIE
 
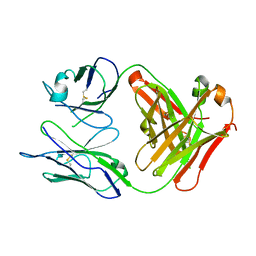 | | Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 | | Descriptor: | IMMUNOGLOBULIN MS5-393 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-23 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
7WKZ
 
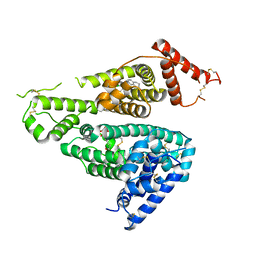 | | Crystal structure of the HSA complex with mycophenolate and aripiprazole | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, MYCOPHENOLIC ACID, Serum albumin | | Authors: | Kawai, A, Yamasaki, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Structural Basis of the Change in the Interaction Between Mycophenolic Acid and Subdomain IIA of Human Serum Albumin During Renal Failure.
J.Med.Chem., 66, 2023
|
|
1MJ7
 
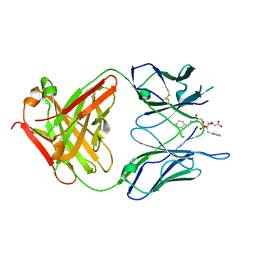 | | Crystal Structure Of The Complex Of The Fab fragment of Esterolytic Antibody MS5-393 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS5-393, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
7Y51
 
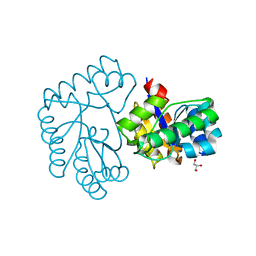 | | Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis TTE0866 delta100 mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Predicted xylanase/chitin deacetylase | | Authors: | Sasamoto, K, Himiyama, T, Moriyoshi, K, Ohmoto, T, Uegaki, K, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Febs Open Bio, 12, 2022
|
|
5XFV
 
 | | Crystal structures of FMN-bound form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
4P7X
 
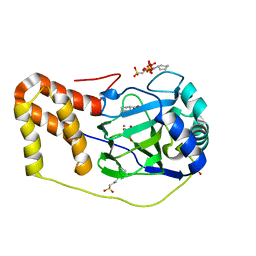 | | L-pipecolic acid-bound L-proline cis-4-hydroxylase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-OXOGLUTARIC ACID, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
4P7W
 
 | | L-proline-bound L-proline cis-4-hydroxylase | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, L-proline cis-4-hydroxylase, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
