1UD4
 
 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD2
 
 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | Descriptor: | GLYCEROL, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
3X0V
 
 | | Structure of L-lysine oxidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-lysine oxidase | | Authors: | Sano, T, Uchida, Y, Amano, M, Kawaguchi, T, Kondo, H, Inagaki, K, Imada, K. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, l-lysine alpha-oxidase from Trichoderma viride.
J.Biochem., 157, 2015
|
|
2D4V
 
 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | Descriptor: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | Authors: | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
1MC8
 
 | | Crystal Structure of Flap Endonuclease-1 R42E mutant from Pyrococcus horikoshii | | Descriptor: | Flap Endonuclease-1 | | Authors: | Matsui, E, Musti, K.V, Abe, J, Yamazaki, K, Matsui, I, Harata, K. | | Deposit date: | 2002-08-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Structure and Novel DNA Binding Sites Located in Loops of Flap Endonuclease-1 from Pyrococcus horikoshii
J.BIOL.CHEM., 277, 2002
|
|
2D4W
 
 | |
1IPA
 
 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2DSK
 
 | | Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, chitinase | | Authors: | Nakamura, T, Mine, S, Hagihara, Y, Ishikawa, K, Uegaki, K. | | Deposit date: | 2006-06-30 | | Release date: | 2007-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the catalytic domain of the hyperthermophilic chitinase from Pyrococcus furiosus
ACTA CRYSTALLOGR.,SECT.F, 63, 2007
|
|
1DUM
 
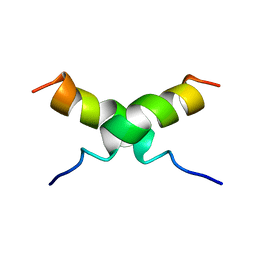 | | NMR STRUCTURE OF [F5Y, F16W] MAGAININ 2 BOUND TO PHOSPHOLIPID VESICLES | | Descriptor: | MAGAININ 2 | | Authors: | Takeda, A, Wakamatsu, K, Tachi, T, Matsuzaki, K. | | Deposit date: | 2000-01-18 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Effects of peptide dimerization on pore formation: Antiparallel disulfide-dimerized magainin 2 analogue.
Biopolymers, 58, 2001
|
|
3AK1
 
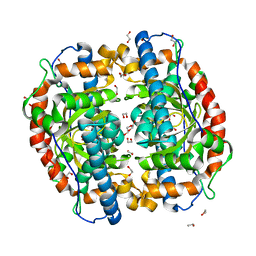 | | Superoxide dismutase from Aeropyrum pernix K1, apo-form | | Descriptor: | 1,2-ETHANEDIOL, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
2E1M
 
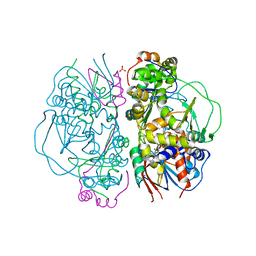 | | Crystal Structure of L-Glutamate Oxidase from Streptomyces sp. X-119-6 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, PHOSPHATE ION | | Authors: | Sasaki, C, Kashima, A, Sakaguchi, C, Mizuno, H, Arima, J, Kusakabe, H, Tamura, T, Sugio, S, Inagaki, K. | | Deposit date: | 2006-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6
Febs J., 276, 2009
|
|
2ZB5
 
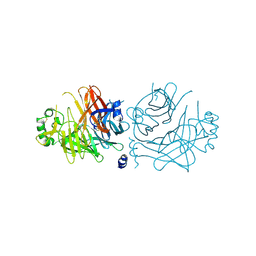 | | Crystal structure of the measles virus hemagglutinin (complex-sugar-type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2GCC
 
 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
2ZB6
 
 | | Crystal structure of the measles virus hemagglutinin (oligo-sugar type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3A8U
 
 | |
3AK3
 
 | | Superoxide dismutase from Aeropyrum pernix K1, Fe-bound form | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
1QQI
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
3AK2
 
 | | Superoxide dismutase from Aeropyrum pernix K1, Mn-bound form | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
2K2G
 
 | | Solution structure of the wild-type catalytic domain of human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor | | Descriptor: | Macrophage metalloelastase, N-(dibenzo[b,d]thiophen-3-ylsulfonyl)-L-valine, ZINC ION | | Authors: | Markus, M.A, Dwyer, B, Wolfrom, S, Li, J, Li, W, Malakian, K, Wilhelm, J, Tsao, D.H.H. | | Deposit date: | 2008-04-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of wild-type human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor.
J.Biomol.Nmr, 41, 2008
|
|
3VP8
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p | | Descriptor: | General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
1IPJ
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS COMPLEXES WITH N-ACETYL-D-GLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
1IPK
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
3A4X
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
