4WR5
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WR4
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
3WOF
 
 | | Crystal structure of P23-45 gp39 (6-132) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | Descriptor: | F-box only protein 43, ZINC ION | | Authors: | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
3WOE
 
 | | Crystal structure of P23-45 gp39 (6-109) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
3WOD
 
 | | RNA polymerase-gp39 complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
2CWX
 
 | | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-1 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-1 crystal)
To be Published
|
|
2D69
 
 | | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase, SULFATE ION | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
2DBR
 
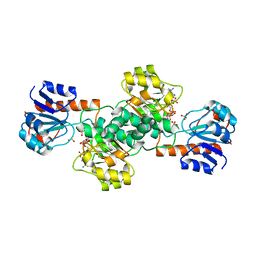 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P1) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBQ
 
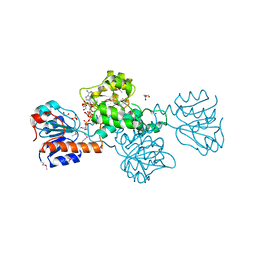 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (I41) | | Descriptor: | GLYCEROL, Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBZ
 
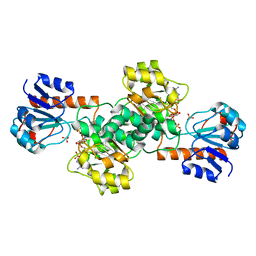 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P61) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBS
 
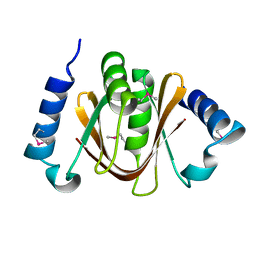 | | Crystal structure of a hypothetical protein TTHC002 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHC002 | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Fusatomi, E, Akasaka, R, Bessho, Y, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a hypothetical protein TTHC002 from Thermus thermophilus HB8
To be Published
|
|
2CXE
 
 | | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
2D54
 
 | | Crystal Structure of Methionyl tRNA Synthetase Y225A Mutant from Thermus Thermophilus | | Descriptor: | Methionyl-tRNA synthetase, ZINC ION | | Authors: | Konno, M, Takeda, R, Takasaka, R, Mori, Y, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-28 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Y225F/Amutation for Met-tRNA synthetase reveals importance of hydrophobic circumstance
To be Published
|
|
2D5B
 
 | | Crystal Structure of Thermus Thermophilus Methionyl tRNA synthetase Y225F Mutant obtained in the presence of PEG6000 | | Descriptor: | Methionyl-tRNA Synthetase, ZINC ION | | Authors: | Konno, M, Takeda, R, Takasaka, R, Mori, Y, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Y225F/A Mutation for Met-tRNA synthetase reveals importance of hydrophobic circumstances
To be Published
|
|
