4FBY
 
 | | fs X-ray diffraction of Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (6.56 Å) | | Cite: | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OW3
 
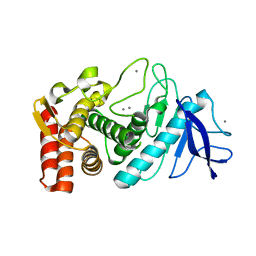 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
3J40
 
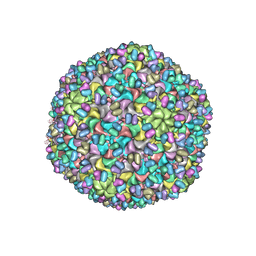 | | Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling | | Descriptor: | gp10, gp7 | | Authors: | Baker, M.L, Hryc, C.F, Zhang, Q, Wu, W, Jakana, J, Haase-Pettingell, C, Afonine, P.V, Adams, P.D, King, J.A, Jiang, W, Chiu, W. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J7N
 
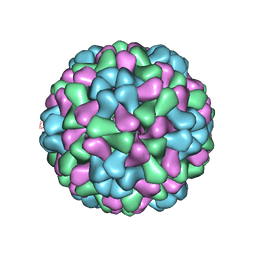 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7L
 
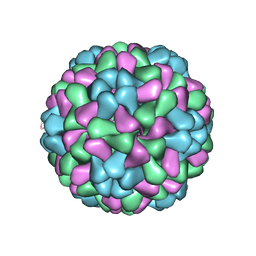 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
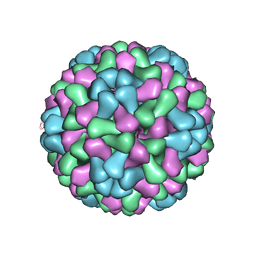 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3OS9
 
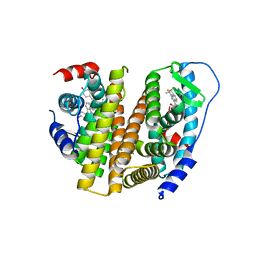 | | Estrogen Receptor | | Descriptor: | 4-[1-allyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
3OS8
 
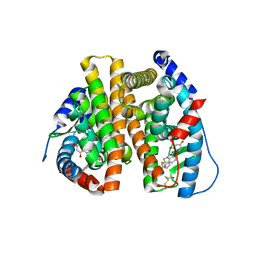 | | Estrogen Receptor | | Descriptor: | 4-[1-benzyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
3OSA
 
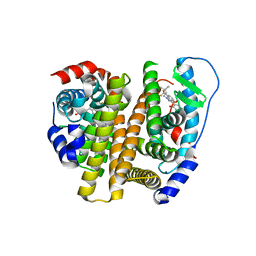 | | Estrogen Receptor | | Descriptor: | 4-[1-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
3KFB
 
 | | Crystal structure of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3KFE
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3PFX
 
 | | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiohydrolase 1 catalytic domain, SULFATE ION, ... | | Authors: | Qin, L, Pereira, J.H, McAndrew, R.P, Simmons, B.A, Sapra, R, Adams, P.D, Sale, K.L. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2616 Å) | | Cite: | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellobiose
TO BE PUBLISHED
|
|
3PFZ
 
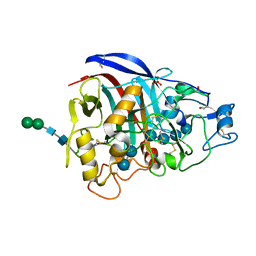 | | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiohydrolase 1 catalytic domain, SULFATE ION, ... | | Authors: | Qin, L, Pereira, J.H, McAndrew, R.P, Simmons, B.A, Sapra, R, Adams, P.D, Sale, K.L. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.0993 Å) | | Cite: | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellotetraose
TO BE PUBLISHED
|
|
3SDR
 
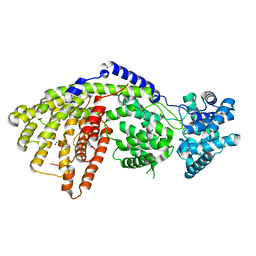 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | Alpha-bisabolene synthase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3SDT
 
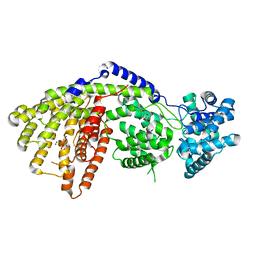 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Alpha-bisabolene synthase, CHLORIDE ION, ... | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3SDV
 
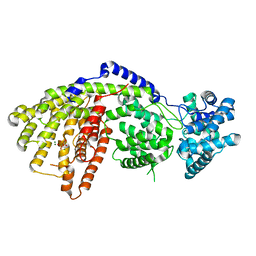 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), Alpha-bisabolene synthase, CHLORIDE ION, ... | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3SAE
 
 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | Alpha-bisabolene synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3SDQ
 
 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | Alpha-bisabolene synthase, CHLORIDE ION, GLYCEROL | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3SDU
 
 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | Alpha-bisabolene synthase, CHLORIDE ION, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3TX8
 
 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RUW
 
 | | Crystal structure of Cpn-rls in complex with ADP-AlFx from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Chaperonin, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUV
 
 | | Crystal structure of Cpn-rls in complex with ATP analogue from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUQ
 
 | | Crystal structure of Cpn-WT in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUS
 
 | | Crystal structure of Cpn-rls in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
4NBT
 
 | | Crystal structure of FabG from Acholeplasma laidlawii | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
