7OXO
 
 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
9F7V
 
 | |
9F7O
 
 | |
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
1EXS
 
 | | STRUCTURE OF PORCINE BETA-LACTOGLOBULIN | | Descriptor: | BETA-LACTOGLOBULIN, GLYCEROL, SODIUM ION | | Authors: | Abrahams, J.P, Hoedemaeker, F.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A novel pH-dependent dimerization motif in beta-lactoglobulin from pig (Sus scrofa).
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1PSI
 
 | |
1UZ8
 
 | | anti-Lewis X Fab fragment in complex with Lewis X | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
1UZ6
 
 | | anti-Lewis X Fab fragment uncomplexed | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
6T17
 
 | |
2BJJ
 
 | | Structure of recombinant human lactoferrin produced in the milk of transgenic cows | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Thomassen, E.A.J, Van Veen, H.A, Van Berkel, P.H.C, Nuijens, J.H, Abrahams, J.P. | | Deposit date: | 2005-02-03 | | Release date: | 2005-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Protein Structure of Recombinant Human Lactoferrin Produced in the Milk of Transgenic Cows Closely Matches the Structure of Human Milk-Derived Lactoferrin
Transgenic Res., 14, 2005
|
|
5O4W
 
 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5O4X
 
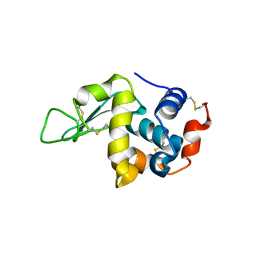 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
9EOL
 
 | |
1COW
 
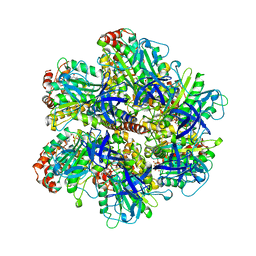 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1MO7
 
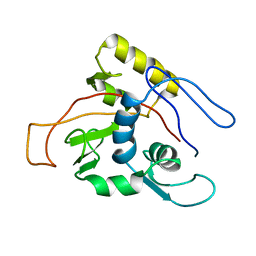 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO8
 
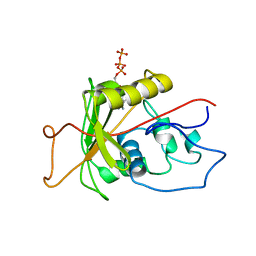 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1SKY
 
 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ6
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6ZHJ
 
 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHN
 
 | | 3D electron diffraction structure of thaumatin from Thaumatococcus daniellii | | Descriptor: | CHLORIDE ION, Thaumatin-1 | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.76 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHB
 
 | | 3D electron diffraction structure of bovine insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
