1WT5
 
 | | The Crystal Structure Of A Humanized Antibody Fv 528 | | Descriptor: | ANTI EGFR ANTIBODY FV REGION | | Authors: | Makabe, K, Tsumoto, K, Asano, R, Kondo, H, Kumagai, I. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic consequences of mutations in vernier zone residues of a humanized anti-human epidermal growth factor receptor murine antibody, 528
J.Biol.Chem., 283, 2008
|
|
3B0O
 
 | | Crystal structure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION | | Authors: | Makabe, K, Kuwajima, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-13 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the stability perturbations induced by N-terminal variation in human and goat alpha-lactalbumin
Protein Eng.Des.Sel., 26, 2013
|
|
3B0I
 
 | | Crystal structure of recombinant human alpha lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION, SULFATE ION | | Authors: | Makabe, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the stability perturbations induced by N-terminal variation in human and goat alpha-lactalbumin
Protein Eng.Des.Sel., 26, 2013
|
|
3B0K
 
 | | Crystal structure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION | | Authors: | Makabe, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the stability perturbations induced by N-terminal variation in human and goat alpha-lactalbumin
Protein Eng.Des.Sel., 26, 2013
|
|
1DG1
 
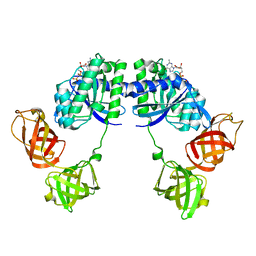 | | WHOLE, UNMODIFIED, EF-TU(ELONGATION FACTOR TU). | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Abel, K, Yoder, M, Hilgenfeld, R, Jurnak, F. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alpha to beta conformational switch in EF-Tu.
Structure, 4, 1996
|
|
1R2J
 
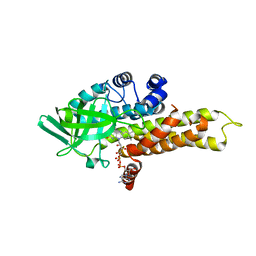 | | FkbI for Biosynthesis of Methoxymalonyl Extender Unit of Fk520 Polyketide Immunosuppresant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, protein FkbI | | Authors: | Watanabe, K, Khosla, C, Stroud, R.M, Tsai, S.-C. | | Deposit date: | 2003-09-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an Acyl-ACP Dehydrogenase from the FK520 Polyketide Biosynthetic Pathway: Insights into Extender Unit Biosynthesis
J.Mol.Biol., 334, 2003
|
|
1V6A
 
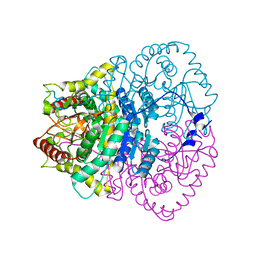 | |
3AUM
 
 | | Crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aromatic cluster mutations produce focal modulations of beta-sheet structure.
Protein Sci., 24, 2015
|
|
1UOK
 
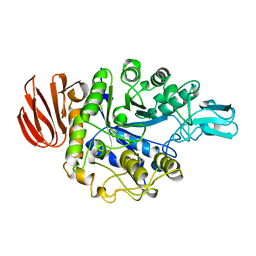 | | CRYSTAL STRUCTURE OF B. CEREUS OLIGO-1,6-GLUCOSIDASE | | Descriptor: | OLIGO-1,6-GLUCOSIDASE | | Authors: | Watanabe, K, Hata, Y, Kizaki, H, Katsube, Y, Suzuki, Y. | | Deposit date: | 1998-07-28 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 A resolution: structural characterization of proline-substitution sites for protein thermostabilization.
J.Mol.Biol., 269, 1997
|
|
3CKG
 
 | | The crystal structure of OspA deletion mutant | | Descriptor: | MAGNESIUM ION, Outer surface protein A | | Authors: | Makabe, K, Koide, S. | | Deposit date: | 2008-03-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The promiscuity of beta-strand pairing allows for rational design of beta-sheet face inversion
J.Am.Chem.Soc., 130, 2008
|
|
3EEX
 
 | | The crystal structure of OspA mutant | | Descriptor: | HEXAETHYLENE GLYCOL, Outer Surface Protein A | | Authors: | Makabe, K, Biancalana, M, Koide, S. | | Deposit date: | 2008-09-06 | | Release date: | 2009-09-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Minimalist design of water-soluble cross-{beta} architecture.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3CKA
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A, PENTAETHYLENE GLYCOL | | Authors: | Makabe, K, Biancalana, M, Terechko, V, Koide, S. | | Deposit date: | 2008-03-14 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Minimalist design of water-soluble cross-{beta} architecture.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3CH8
 
 | | The crystal structure of PDZ-Fibronectin fusion protein | | Descriptor: | C-terminal octapeptide from protein ARVCF, MAGNESIUM ION, fusion protein PDZ-Fibronectin,Fibronectin | | Authors: | Makabe, K, Huang, J, Koide, A, Koide, S. | | Deposit date: | 2008-03-08 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution.
J.Mol.Biol., 392, 2009
|
|
3CKF
 
 | | The crystal structure of OspA deletion mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Koide, S. | | Deposit date: | 2008-03-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The promiscuity of beta-strand pairing allows for rational design of beta-sheet face inversion
J.Am.Chem.Soc., 130, 2008
|
|
1J1R
 
 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Antiviral Protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1Q
 
 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1GP9
 
 | | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Watanabe, K, Chirgadze, D.Y, Lietha, D, Gherardi, E, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping
J.Mol.Biol., 319, 2002
|
|
1J1S
 
 | | Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Formycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral Protein S, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1M
 
 | | Ricin A-Chain (Recombinant) at 100K | | Descriptor: | Ricin, SULFATE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Watanabe, K, Motoshima, H. | | Deposit date: | 2002-12-10 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ricin A-Chain (Recombinant) at 100K
To be published
|
|
7X2B
 
 | | Red fluorescent protein from Diadumene lineata | | Descriptor: | Red fluorescent protein, SULFATE ION | | Authors: | Makabe, K, Hotta, J, Mizuno, H. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Cloning and structural basis of fluorescent protein color variants from identical species of sea anemone, Diadumene lineata.
Photochem Photobiol Sci, 22, 2023
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
3W1F
 
 | | Crystal structure of Human MPS1 catalytic domain in complex with 5-(5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl)-2-methylbenzenesulfonamide | | Descriptor: | 5-[5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl]-2-methylbenzenesulfonamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Tachibana, Y, Itoh, T, Yamamoto, T, Hashizume, H, Hato, Y, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Higaki, M, Ueda, K, Yoshizawa, H, Baba, Y, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-11-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Indazole-based potent and cell-active Mps1 kinase inhibitors: rational design from pan-kinase inhibitor anthrapyrazolone (SP600125)
J.Med.Chem., 56, 2013
|
|
2Z3K
 
 | | complex structure of LF-transferase and rAF | | Descriptor: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, ... | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
2Z3M
 
 | | complex structure of LF-transferase and dAF | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, ... | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
2Z3L
 
 | | complex structure of LF-transferase and peptide A | | Descriptor: | D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, peptide (PHE)(ARG)(TYR)(LEU)(GLY) | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
