2OY7
 
 | |
2OYB
 
 | |
3B0K
 
 | | Crystal structure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION | | Authors: | Makabe, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the stability perturbations induced by N-terminal variation in human and goat alpha-lactalbumin
Protein Eng.Des.Sel., 26, 2013
|
|
3B0O
 
 | | Crystal structure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION | | Authors: | Makabe, K, Kuwajima, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the stability perturbations induced by N-terminal variation in human and goat alpha-lactalbumin
Protein Eng.Des.Sel., 26, 2013
|
|
3B0I
 
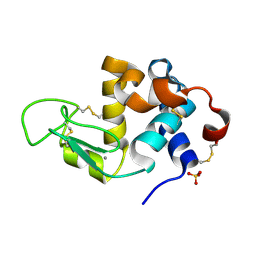 | | Crystal structure of recombinant human alpha lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION, SULFATE ION | | Authors: | Makabe, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the stability perturbations induced by N-terminal variation in human and goat alpha-lactalbumin
Protein Eng.Des.Sel., 26, 2013
|
|
3AUM
 
 | | Crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aromatic cluster mutations produce focal modulations of beta-sheet structure.
Protein Sci., 24, 2015
|
|
2FKJ
 
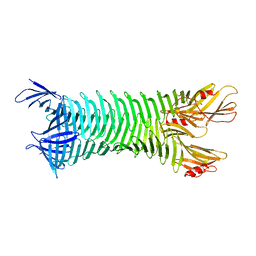 | | The crystal structure of engineered OspA | | Descriptor: | Outer Surface Protein A | | Authors: | Makabe, K, Terechko, V, Gawlak, G, Yan, S, Koide, S. | | Deposit date: | 2006-01-04 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6M8N
 
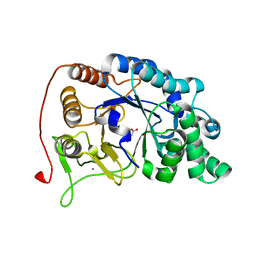 | | Endo-fucoidan hydrolase P5AFcnA from glycoside hydrolase family 107 | | Descriptor: | CALCIUM ION, MALONATE ION, P5AFcnA | | Authors: | Boraston, A.B, Vickers, C.J, Abe, K, Salama-Alber, O. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
1EQK
 
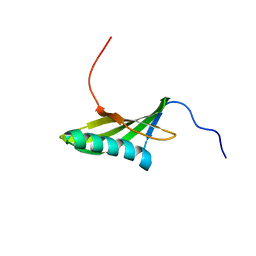 | | SOLUTION STRUCTURE OF ORYZACYSTATIN-I, A CYSTEINE PROTEINASE INHIBITOR OF THE RICE, ORYZA SATIVA L. JAPONICA | | Descriptor: | ORYZACYSTATIN-I | | Authors: | Nagata, K, Kudo, N, Abe, K, Arai, S, Tanokura, M. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica.
Biochemistry, 39, 2000
|
|
6DLH
 
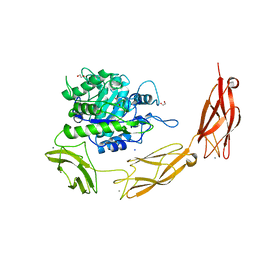 | | Endo-fucoidan hydrolase MfFcnA4 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION, ... | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
6DMS
 
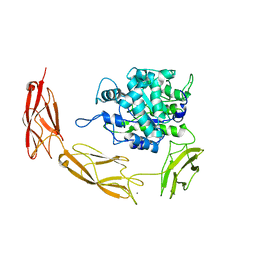 | | Endo-fucoidan hydrolase MfFcnA4_H294Q from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
4UAP
 
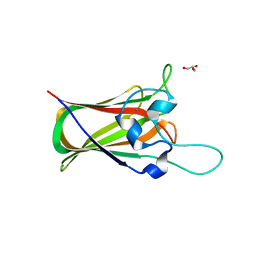 | | X-ray structure of GH31 CBM32-2 bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Grondin, J.M, Abe, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
6DNS
 
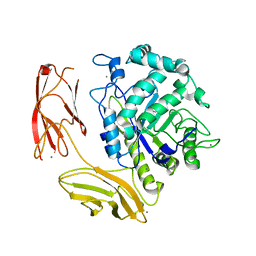 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
7X22
 
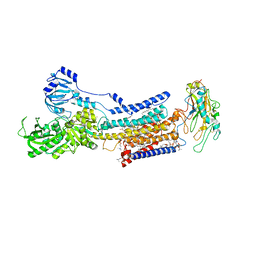 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794S in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7X21
 
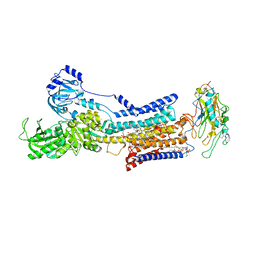 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
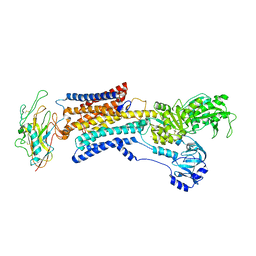
7X20
 
 | |
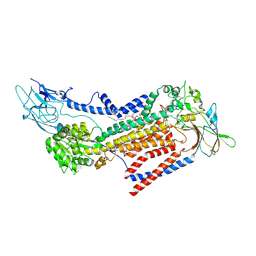
7VSG
 
 | |
7VSH
 
 | |
5FQ0
 
 | | The structure of KdgF from Halomonas sp. | | Descriptor: | CITRATE ANION, KDGF, NICKEL (II) ION, ... | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FPZ
 
 | | The structure of KdgF from Yersinia enterocolitica with malonate bound in the active site. | | Descriptor: | MALONIC ACID, NICKEL (II) ION, PECTIN DEGRADATION PROTEIN | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FPX
 
 | | The structure of KdgF from Yersinia enterocolitica. | | Descriptor: | NICKEL (II) ION, PECTIN DEGRADATION PROTEIN, PEPTIDE | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FQE
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQF
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
4ZO9
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZO6
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with cellobiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
