4OQL
 
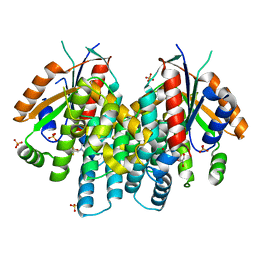 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with dF-EdU | | Descriptor: | 2'-deoxy-5-ethynyl-2',2'-difluorouridine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with dF-EdU
To be Published
|
|
4OQM
 
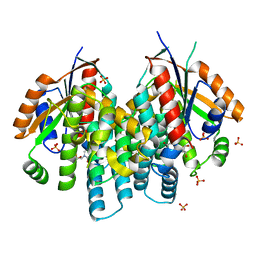 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU | | Descriptor: | 1-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)-5-ethynylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU
To be Published
|
|
4OTA
 
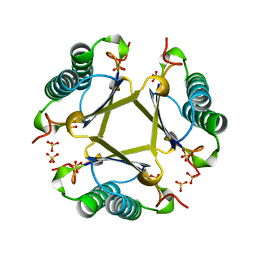 | |
8JSH
 
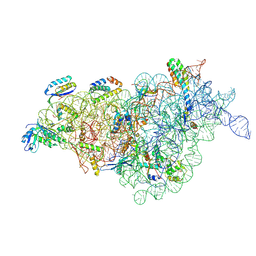 | |
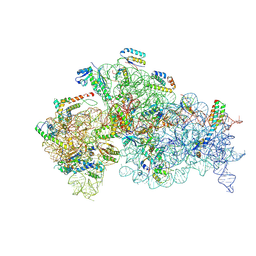
8JSG
 
 | |
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
2MCG
 
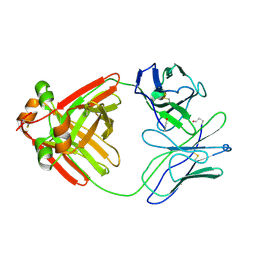 | |
1UB5
 
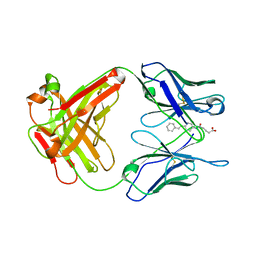 | | Crystal structure of Antibody 19G2 with hapten at 100K | | Descriptor: | 4-(4-STYRYL-PHENYLCARBAMOYL)-BUTYRIC ACID, antibody 19G2, alpha chain, ... | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
4D1Q
 
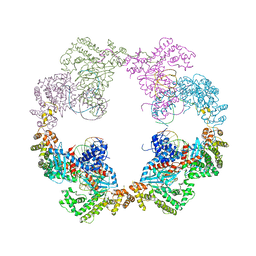 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
1TTK
 
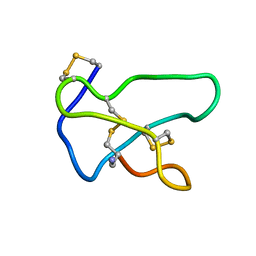 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TT3
 
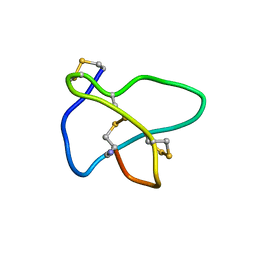 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1U8E
 
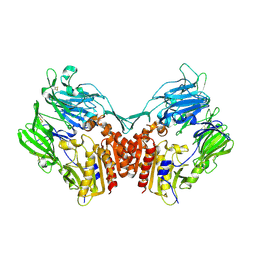 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
4E27
 
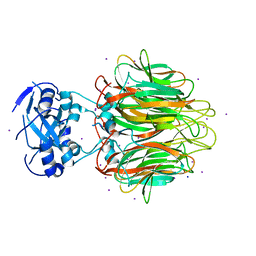 | | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences Solved by Iodide SAD Phasing | | Descriptor: | Capsid Protein, IODIDE ION, SODIUM ION | | Authors: | Craig, T.K, Abendroth, J, Lorimer, D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences Solved by Iodide SAD Phasing
To be Published
|
|
1TU6
 
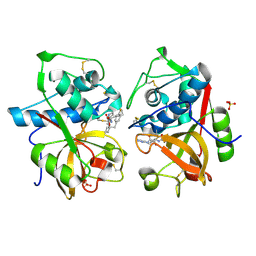 | | Cathepsin K complexed with a ketoamide inhibitor | | Descriptor: | Cathepsin K, SULFATE ION, [1-(4-FLUOROBENZYL)CYCLOBUTYL]METHYL (1S)-1-[OXO(1H-PYRAZOL-5-YLAMINO)ACETYL]PENTYLCARBAMATE | | Authors: | Barrett, D.G, Catalano, J.G, Deaton, D.N, Hassell, A.M, Long, S.T, Miller, A.B, Miller, L.R, Shewchuk, L.M, Wells-Knecht, K.J, Wright, L.L. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent and selective P2-P3 ketoamide inhibitors of cathepsin K with improved pharmacokinetic properties via favorable P1', P1, and/or P3 substitutions
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
1UB6
 
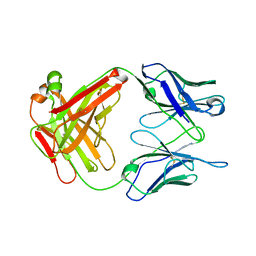 | | Crystal structure of Antibody 19G2 with sera ligand | | Descriptor: | antibody 19G2, alpha chain, beta chain | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
6QTF
 
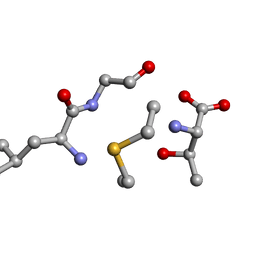 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, major conformer | | Descriptor: | DCY-LEU-GLY-ALA-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYU
 
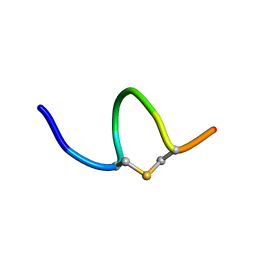 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A | | Descriptor: | PHE-DHA-DAL-LEU-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYR
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, minor conformer | | Descriptor: | DAL-LEU-GLY-CYS-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QM1
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B (Lan8,11) analogue | | Descriptor: | DAL-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-01 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6SES
 
 | | Tubulin-B2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Guo, B, Rodriguez-Gabin, A, Prota, A.E, Muehlethaler, T, Zhang, N, Ye, K, Steinmetz, M.O, Band Horwitz, S, Smith III, A.B, McDaid, H.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence.
Mol.Pharmacol., 98, 2020
|
|
2WCS
 
 | | Crystal Structure of Debranching enzyme from Nostoc punctiforme (NPDE) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.B, Choi, J.H, Nam, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
2W8F
 
 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
2WKG
 
 | | Nostoc punctiforme Debranching Enzyme (NPDE)(Native form) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.B, Choi, J.H, Song, H.N, Park, K.H, Woo, E.J. | | Deposit date: | 2009-06-11 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
2YA1
 
 | | Product complex of a multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | PUTATIVE ALKALINE AMYLOPULLULANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
