1IGM
 
 | |
1UY4
 
 | |
1UY2
 
 | |
1UY3
 
 | |
1UY1
 
 | |
1YK8
 
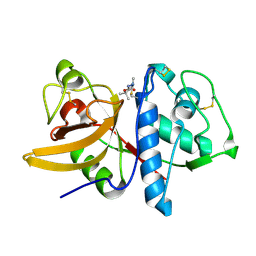 | | Cathepsin K complexed with a cyanamide-based inhibitor | | Descriptor: | Cathepsin K, TERT-BUTYL 2-CYANO-2-METHYLHYDRAZINECARBOXYLATE | | Authors: | Barrett, D.G, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Shewchuk, L.M, Willard, D.H, Wright, L.L. | | Deposit date: | 2005-01-17 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acyclic cyanamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YK7
 
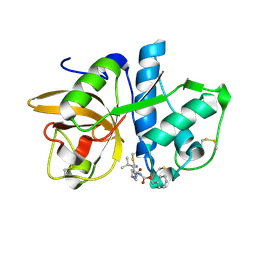 | | Cathepsin K complexed with a cyanopyrrolidine inhibitor | | Descriptor: | Cathepsin K, N2-[(BENZYLOXY)CARBONYL]-N1-[(3S)-1-CYANOPYRROLIDIN-3-YL]-L-LEUCINAMIDE | | Authors: | Barrett, D.G, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Tavares, F.X, Willard, D.H, Wright, L.L. | | Deposit date: | 2005-01-17 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel and potent cyclic cyanamide-based cathepsin K inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YT7
 
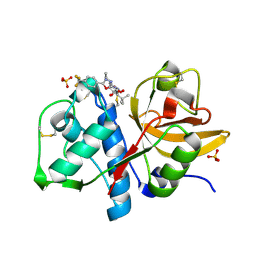 | | Cathepsin K complexed with a constrained ketoamide inhibitor | | Descriptor: | (1R)-2,2-DIMETHYL-1-({5-[4-(TRIFLUOROMETHYL)PHENYL]-1,3,4-OXADIAZOL-2-YL}METHYL)PROPYL (1S)-1-{OXO[(2-OXO-1,3-OXAZOLIDIN-3-YL)AMINO]ACETYL}PENTYLCARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Barrett, D.G, Boncek, V.M, Catalano, J.G, Deaton, D.N, Hassell, A.M, Jurgensen, C.H, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Payne, J.A, Ray, J.A, Samano, V, Shewchuk, L.M, Tavares, F.X, Wells-Knecht, K.J, Willard, D.H, Wright, L.L, Zhou, H.Q. | | Deposit date: | 2005-02-10 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | P(2)-P(3) conformationally constrained ketoamide-based inhibitors of cathepsin K.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1JVK
 
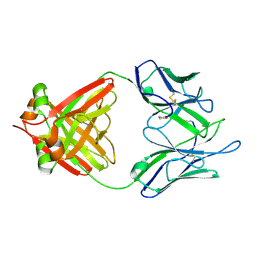 | | THREE-DIMENSIONAL STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN DIMER ACTING AS A LETHAL AMYLOID PRECURSOR | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Bourne, P.C, Ramsland, P.A, Shan, L, Fan, Z.-C, DeWitt, C.R, Shultz, B.B, Terzyan, S.S, Edmundson, A.B. | | Deposit date: | 2001-08-30 | | Release date: | 2002-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2O84
 
 | | Crystal structure of K206E mutant of N-lobe human transferrin | | Descriptor: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | Authors: | Baker, H.M, Nurizzo, D, Mason, A.B, Baker, E.N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2MCG
 
 | |
1JQF
 
 | | Human Transferrin N-Lobe Mutant H249Q | | Descriptor: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | Authors: | Baker, H.M, Mason, A.B, He, Q.-Y, MacGillivray, R.T.A, Baker, E.N. | | Deposit date: | 2001-08-06 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ligand variation in the transferrin family: the crystal structure of the H249Q mutant of the human transferrin N-lobe as a model for iron binding in insect transferrins.
Biochemistry, 40, 2001
|
|
2LLW
 
 | | Solution structure of the yeast Sti1 DP2 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
2M6T
 
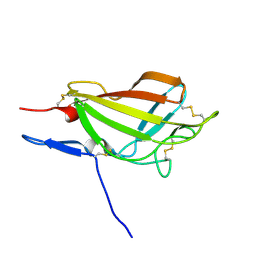 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-04-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2O7U
 
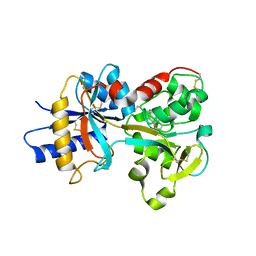 | | Crystal structure of K206E/K296E mutant of the N-terminal half molecule of human transferrin | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Baker, H.M, Nurizzo, D, Mason, A.B, Baker, E.N. | | Deposit date: | 2006-12-11 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2L21
 
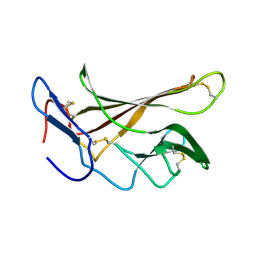 | | chicken IGF2R domain 11 | | Descriptor: | Cation-independent mannose-6-phosphate receptor | | Authors: | Williams, C, Hoppe, H, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Rezgui, D.Z, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | CD loop dependency of the IGF2: M6P/IGF2 receptor binding interaction predates imprinting
To be Published
|
|
2L29
 
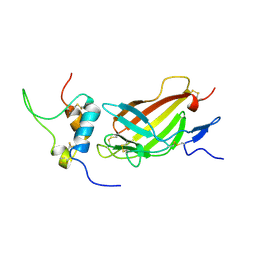 | | Complex structure of E4 mutant human IGF2R domain 11 bound to IGF-II | | Descriptor: | Insulin-like growth factor 2 receptor variant, Insulin-like growth factor II | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
2LM3
 
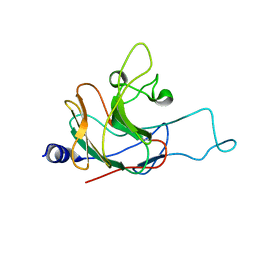 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2O4E
 
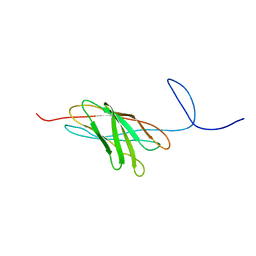 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
3KBE
 
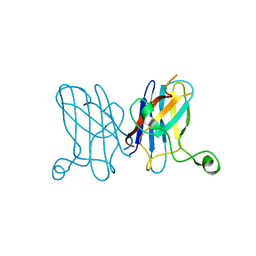 | | Metal-free C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
4ECC
 
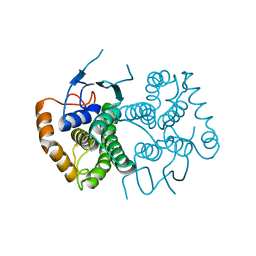 | | Chimeric GST Containing Inserts of Kininogen Peptides | | Descriptor: | chimeric protein between GSHKT10 and domain 5 of kininogen-1 | | Authors: | Amber, A.B, Sergei, M.M, Yi, P, Rita, R, Xiaoping, Q, Marianne, P.-C, William, C.M, Vivien, Y, Keith, R.M, Anton, A.K. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric glutathione S-transferases containing inserts of kininogen peptides: potential novel protein therapeutics.
J.Biol.Chem., 287, 2012
|
|
3KN7
 
 | |
3KBF
 
 | | C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (II) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
3KM1
 
 | | ZINC-Reconstituted TOMATO CHLOROPLAST SUPEROXIDE DISMUTASE | | Descriptor: | GLYCEROL, Superoxide dismutase [Cu-Zn], chloroplastic, ... | | Authors: | Galaleldeen, A, Taylor, A.B, Thomas, S.T, Hart, P.J. | | Deposit date: | 2009-11-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biophysical properties of tomato chloroplast sod
To be Published
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
