3JBW
 
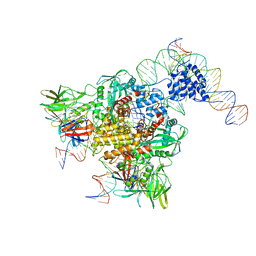 | | Cryo-electron microscopy structure of RAG Paired Complex (with NBD, no symmetry) | | Descriptor: | 12-RSS signal end forward strand, 5'-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', Nicked 12-RSS intermediate reverse strand, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3KBL
 
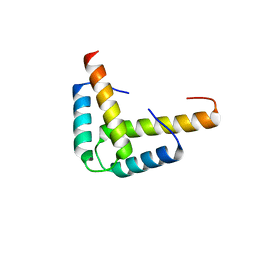 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
7JW4
 
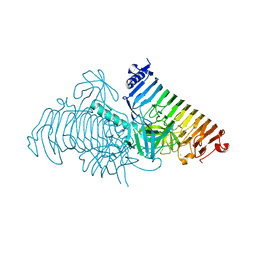 | | Crystal structure of PdGH110B in complex with D-galactose | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 110, NICKEL (II) ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | The structure of a family 110 glycoside hydrolase provides insight into the hydrolysis of alpha-1,3-galactosidic linkages in lambda-carrageenan and blood group antigens.
J.Biol.Chem., 295, 2020
|
|
5SVB
 
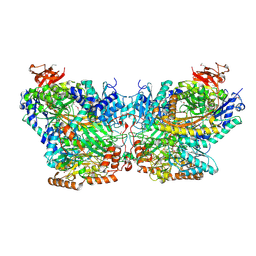 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
7JTV
 
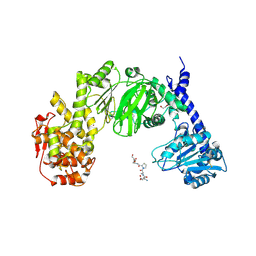 | |
5SWA
 
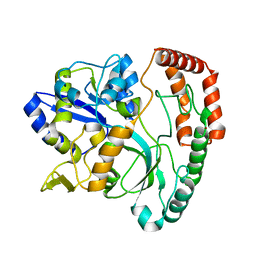 | |
3LZL
 
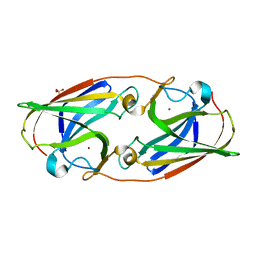 | | Crystal Structure Analysis of the as-solated P19 protein from Campylobacter jejuni at 1.45 A at pH 9.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
3MHS
 
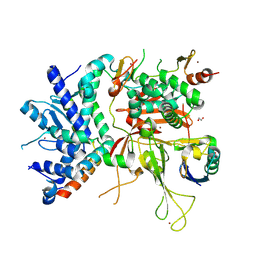 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
3VFI
 
 | | Crystal Structure of a Metagenomic Thioredoxin | | Descriptor: | thioredoxin | | Authors: | Craig, T.K, Gardberg, A, Lorimer, D.D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Metagenomic Thioredoxin
To be Published
|
|
7JRL
 
 | | The structure of CBM51-2 in complex with GlcNAc and INT domains from Clostridium perfringens ZmpB | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-08-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecturally complex O -glycopeptidases are customized for mucin recognition and hydrolysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3KLY
 
 | |
3KLZ
 
 | |
3UAQ
 
 | | Crystal Structure of the N-lobe Domain of Lactoferrin Binding Protein B (LbpB) of Moraxella bovis | | Descriptor: | LbpB B-lobe | | Authors: | Arutyunova, E, Brooks, C.L, Beddeck, A, Mak, M.W, Schryvers, A.B, Lemieux, M.J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9318 Å) | | Cite: | Crystal structure of the N-lobe of lactoferrin binding protein B from Moraxella bovis(1).
Biochem.Cell Biol., 90, 2012
|
|
5T98
 
 | | Crystal structure of BuGH2Awt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA1
 
 | | Crystal structure of BuGH86wt | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA9
 
 | |
3KN8
 
 | |
3MMP
 
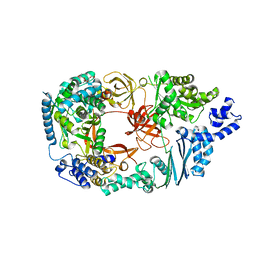 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5T9G
 
 | |
5TA7
 
 | | Crystal structure of BuGH117Bwt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T3U
 
 | |
5SVC
 
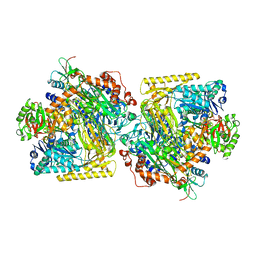 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5SUO
 
 | |
4GDP
 
 | | Yeast polyamine oxidase FMS1, N195A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, TETRAETHYLENE GLYCOL | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9998 Å) | | Cite: | Mechanistic and Structural Analyses of the Roles of Active Site Residues in Yeast Polyamine Oxidase Fms1: Characterization of the N195A and D94N Enzymes.
Biochemistry, 51, 2012
|
|
4ECH
 
 | | Yeast Polyamine Oxidase FMS1, H67Q Mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic and structural analyses of the role of his67 in the yeast polyamine oxidase fms1.
Biochemistry, 51, 2012
|
|
