6ZZA
 
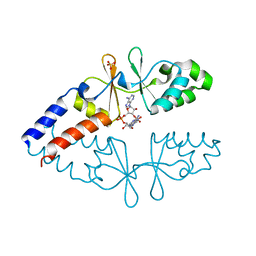 | | Crystal structure of CbpB in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein, SULFATE ION | | Authors: | Mechaly, A.E, Covaleda-Cortes, G, Kaminski, P.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of CbpB in complex with c-di-AMP
To Be Published
|
|
6U0R
 
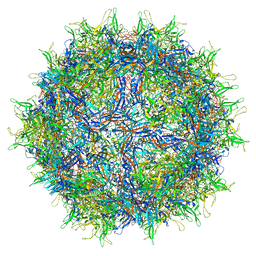 | |
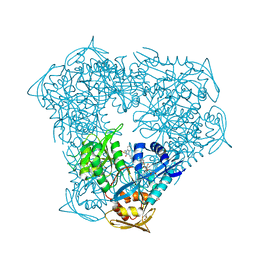
6I4X
 
 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with erythropoietin receptor peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
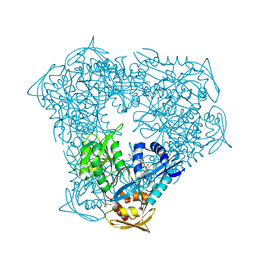
6X6G
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with droperidol | | Descriptor: | 3-[1-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-2,3,4,5-tetrahydropyridin-4-yl]-1~{H}-benzimidazol-2-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
6X6I
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT543 | | Descriptor: | 4-(4-benzyl-4-hydroxypiperidin-1-yl)-1-(4-fluorophenyl)butan-1-one, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
6X5Y
 
 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
2N7K
 
 | | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8 | | Descriptor: | NEDD8, Protein KHNYN | | Authors: | Santonico, E, Nepravishta, R, Mattioni, A, Valentini, E, Mandaliti, W, Procopio, R, Iannuccelli, M, Castagnoli, L, Polo, S, Paci, M, Cesareni, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8
To be Published
|
|
6X6Y
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1264 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
1CZK
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 2000-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
6X7A
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT572 | | Descriptor: | 4-(4-cyclohexyl-3,4-dihydro-2~{H}-pyridin-1-yl)-1-(4-$l^{2}-fluoranylcyclohexa-1,3,5-trien-1-yl)butan-1-one, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
1LMZ
 
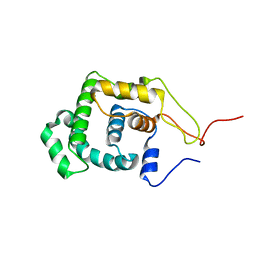 | |
8D2N
 
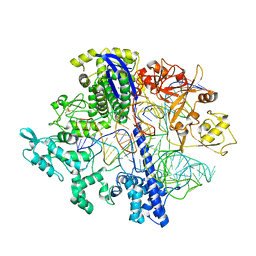 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
1CXI
 
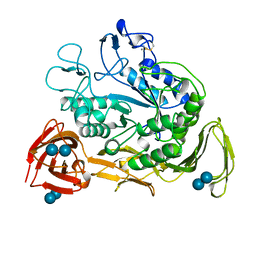 | |
2ELJ
 
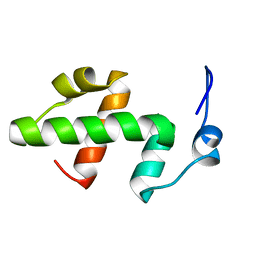 | | Solution structure of the SWIRM domain of baker's yeast Transcriptional adapter 2 | | Descriptor: | Transcriptional adapter 2 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIRM domain of baker's yeast Transcriptional adapter 2
To be Published
|
|
7PEO
 
 | | Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, HOLMIUM ATOM, ... | | Authors: | von Kugelgen, A, Bharat, T.A.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | High-resolution mapping of metal ions reveals principles of surface layer assembly in Caulobacter crescentus cells.
Structure, 30, 2022
|
|
6OT2
 
 | | Structure of the TRPV3 K169A sensitized mutant in apo form at 4.1 A resolution | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
7A2U
 
 | |
7A3D
 
 | |
5I70
 
 | | Crystal Structure of plasmepsin IV | | Descriptor: | Pepstatin A, Plasmepsin IV | | Authors: | Asojo, O.A. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of plasmepsin IV
To Be Published
|
|
2EAN
 
 | | Solution structure of the N-terminal SAM-domain of human KIAA0902 protein (connector enhancer of kinase suppressor of ras 2) | | Descriptor: | Connector enhancer of kinase suppressor of ras 2 | | Authors: | Goroncy, A.K, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-31 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal SAM-domain of human KIAA0902 protein (connector enhancer of kinase suppressor of ras 2)
To be Published
|
|
8AHQ
 
 | | VirD/holo-ACP5b of Streptomyces virginiae complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, CHLORIDE ION, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AHZ
 
 | | Native VirD of Streptomyces virginiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA hydratase, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
7PGM
 
 | | HHIP-C in complex with heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-3)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7AE4
 
 | |
1CQT
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX CONTAINING AN OCA-B PEPTIDE, THE OCT-1 POU DOMAIN, AND AN OCTAMER ELEMENT | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), POU DOMAIN, ... | | Authors: | Chasman, D.I, Cepek, K, Sharp, P.A, Pabo, C.O. | | Deposit date: | 1999-08-11 | | Release date: | 1999-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface.
Genes Dev., 13, 1999
|
|
