8BMZ
 
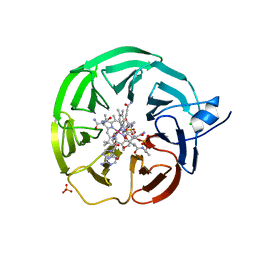 | | Bacteroides thetaiotaomicron surface lipoprotein BT1954 bound to adenosylcobalamin | | Descriptor: | Adenosylcobalamin, CHLORIDE ION, Putative surface layer protein, ... | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
6ONP
 
 | |
7MRC
 
 | |
6ONQ
 
 | |
7A1D
 
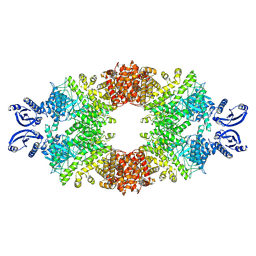 | | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation) | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
7OR0
 
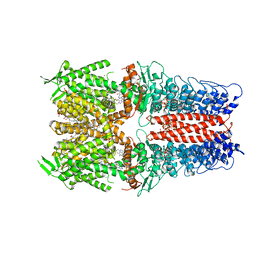 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
6OPM
 
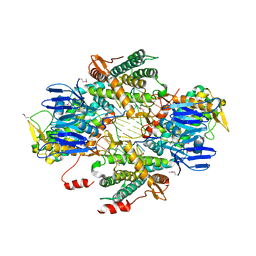 | | Casposase bound to integration product | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, DNA 21-mer, ... | | Authors: | Dyda, F, Hickman, A.B, Kailasan, S. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Elife, 9, 2020
|
|
6UFI
 
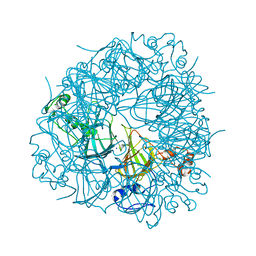 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
6UAQ
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein, SODIUM ION | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
8BMY
 
 | | Bacteroides thetaiotaomicron surface lipoprotein bound to cyanocobalamin | | Descriptor: | CHLORIDE ION, CYANOCOBALAMIN, Putative surface layer protein, ... | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
5IRR
 
 | | Crystal structure of Septin GTPase domain from Chlamydomonas reinhardtii | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Septin-like protein | | Authors: | Pinto, A.P.A, Pereira, H.M, Navarro, M.V.A.S, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Filaments and fingers: Novel structural aspects of the single septin from Chlamydomonas reinhardtii.
J. Biol. Chem., 292, 2017
|
|
6UAW
 
 | | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Pseudomonas viridiflava (PvGH128_II) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, SULFATE ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Santos, C.R, Costa, P.A.C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB2
 
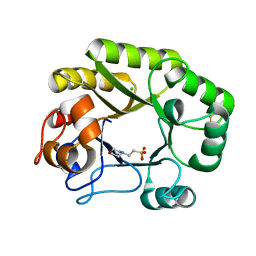 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Endo-beta-1,3-glucanase, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6O5R
 
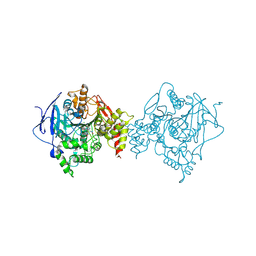 | |
7MVW
 
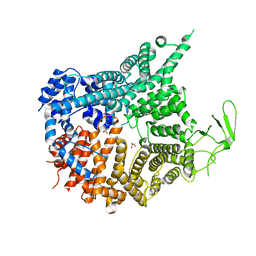 | | Crystal structure of Chaetomium thermophilum Nup188 NTD (residues 1-1134) | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
8BJD
 
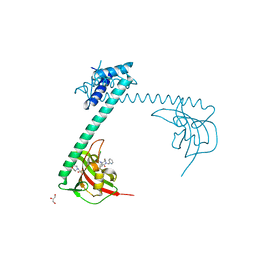 | | Full length structure of LpMIP with bound inhibitor JK095 | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8DNA
 
 | |
8BK4
 
 | | Full length structure of the apo-state LpMIP. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Macrophage infectivity potentiator, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
6WPE
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8E82
 
 | | Mycobacterium tuberculosis RNAP elongation complex with NusG transcription factor | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
6O7N
 
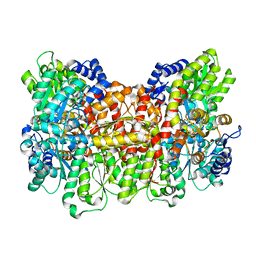 | |
8DN9
 
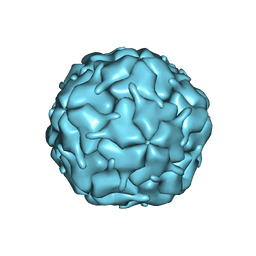 | |
7MVX
 
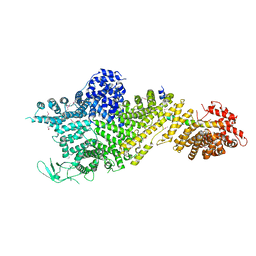 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
8E3N
 
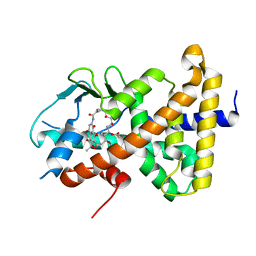 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6MCJ
 
 | |
