4MKT
 
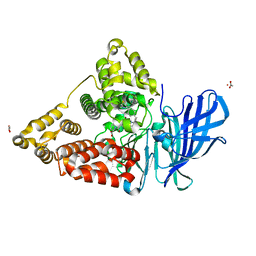 | | Human Leukotriene A4 Hydrolase in complex with Pro-Gly-Pro analogue and 4-(4-benzylphenyl)thiazol-2-amine | | Descriptor: | 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, 4-(4-benzylphenyl)-1,3-thiazol-2-amine, ACETIC ACID, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-09-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8CIE
 
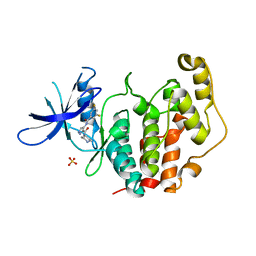 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | Descriptor: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | Authors: | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
5IFA
 
 | |
1KQW
 
 | | Crystal structure of holo-CRBP from zebrafish | | Descriptor: | Cellular retinol-binding protein, RETINOL | | Authors: | Calderone, V, Folli, C, Marchesani, A, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
J.Mol.Biol., 321, 2002
|
|
5IGK
 
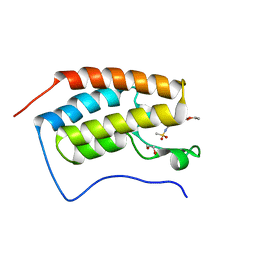 | | Crystal structure of the first bromodomain of human BRD4 in complex with bromosporine (BSP) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
5IED
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1KR7
 
 | | Crystal structure of the nerve tissue mini-hemoglobin from the nemertean worm Cerebratulus lacteus | | Descriptor: | ACETATE ION, Neural globin, OXYGEN MOLECULE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Geuens, E, Yamauchi, k, Ascenzi, P, Riggs, A.F, Moens, L, Bolognesi, M. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 109 residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the alpha-helical globin fold
Structure, 10, 2002
|
|
5IF6
 
 | |
1KRJ
 
 | | Engineering Calcium-binding site into Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Sundaramoorthy, M, Poulos, T.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conversion of an engineered potassium-binding site into a calcium-selective site in cytochrome c peroxidase.
J.Biol.Chem., 274, 1999
|
|
6XXD
 
 | |
5IPK
 
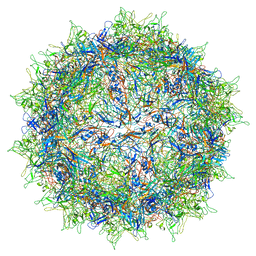 | | Structure of the R432A variant of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
1KS2
 
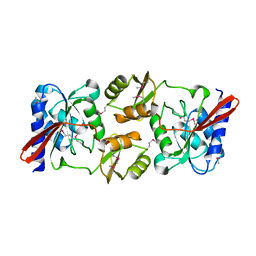 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
5XMR
 
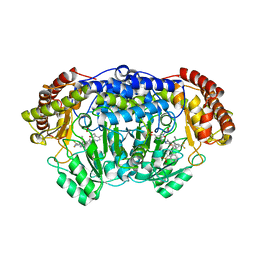 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and GS395 | | Descriptor: | (4~{S})-6-azanyl-3-methyl-4-[3-[4-(phenylmethyl)sulfonylphenyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-2~{H}-pyrano[2 ,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
8COY
 
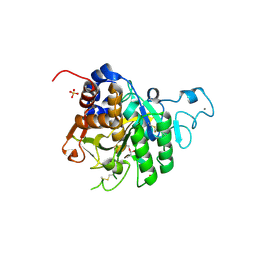 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5IIZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5X5A
 
 | | Human thymidylate synthase bound with phosphate ion | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5IFK
 
 | | Purine nucleoside phosphorylase | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Thakur, K.G, Priyanka, A. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Functional and Structural Characterization of Purine Nucleoside Phosphorylase from Kluyveromyces lactis and Its Potential Applications in Reducing Purine Content in Food
PLoS ONE, 11, 2016
|
|
7NNP
 
 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
1KF3
 
 | | Atomic Resolution Structure of RNase A at pH 5.9 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2P47
 
 | | Complex of a camelid single-domain vhh antibody fragment with RNASE A at 2.5A resolution: SE5B-TRI crystal form with five se-met sites (L4M, M34, M51, F68M, M83) in vhh scaffold. | | Descriptor: | ANTIBODY CAB-RN05, Ribonuclease pancreatic | | Authors: | Tereshko, V, Uysal, S, Koide, A, Margalef, K, Koide, S, Kossiakoff, A.A. | | Deposit date: | 2007-03-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold
Protein Sci., 17, 2008
|
|
1KNM
 
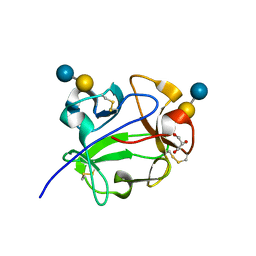 | | Streptomyces lividans Xylan Binding Domain cbm13 in Complex with Lactose | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
7NNL
 
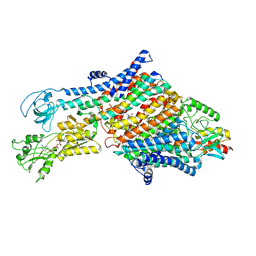 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
8CLR
 
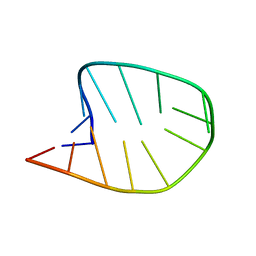 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
1KG3
 
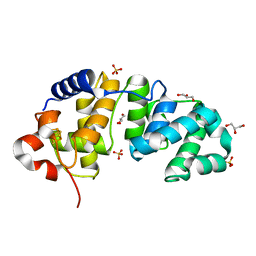 | | Crystal structure of the core fragment of MutY from E.coli at 1.55A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
8CP0
 
 | | Structure of the catalytic domain of P. vivax Sub1 (trigonal crystal form) | | Descriptor: | CALCIUM ION, subtilisin | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
