8VUC
 
 | | Structure of FabS1CE2-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
7ASK
 
 | | 43S preinitiation complex from Trypanosoma cruzi with the kDDX60 helicase bound with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, kDDX60 | | Authors: | Bochler, A, Brito Querido, J, Prilepskaja, T, Soufari, H, Del Cistia, M.L, Kuhn, L, Rimoldi Ribeiro, A, Valasek, L.S, Hashem, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts.
Cell Rep, 33, 2020
|
|
6Y6J
 
 | | VIM-2 in Complex with Biapenem Imine and Enamine Hydrolysis Products | | Descriptor: | Biapenem Enamine hydrolysis product, Biapenem Imine hydrolysis product, CHLORIDE ION, ... | | Authors: | Lucic, A, Schofield, C.J, Brem, J, McDonough, M.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of metallo-beta-lactamase VIM-2 in complex with the imine and enamine form of hydrolysed Biapenem
To Be Published
|
|
8VTP
 
 | | Structure of FabS1CE-EPR-1, a high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
5N6B
 
 | | Human Leukocyte Antigen Class I A02 Carrying LLWNPGMAV | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Bulek, A.M, Bovay, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | T cell receptor alpha variable 12-2 bias in the immunodominant response to Yellow fever virus.
Eur. J. Immunol., 48, 2018
|
|
5N8P
 
 | |
6QDJ
 
 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | Descriptor: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meinhart, A, Clausen, T, Arnese, R. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
7AKM
 
 | | Crystal structure of CHK1 kinase domain in complex with ATPyS | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for recruitment of the CHK1 DNA damage kinase by the CLASPIN scaffold protein.
Structure, 29, 2021
|
|
8VU4
 
 | | Structure of FabS1CE4-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8F51
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with mefloquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
6VZW
 
 | | TTLL6 bound to the initiation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, T.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VI9
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - an alkylperoxo bound structure | | Descriptor: | (5R,6Z)-5-(hydroperoxy-kappaO)-5-(hydroxy-kappaO)-6-iminocyclohexa-1,3-diene-1-carboxylato(2-)iron, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SPE
 
 | | Pseudomonas aeruginosa 30s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8VU1
 
 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (trigonal form) | | Descriptor: | S1CE3 VARIANT OF FAB-EPR-1 heavy chain, S1CE3 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
8VTR
 
 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
6W22
 
 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8EXC
 
 | | Human Carbonic Anhydrase II bound tert-butyl (3-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propoxy)butoxy)propyl)carbamate | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-(3-{4-[3-({2-[(3R)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}amino)propoxy]butoxy}propyl)-4-sulfamoylbenzamide, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8BLO
 
 | | Human Urea Transporter UT-A (N-Terminal Domain Model) | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Urea transporter 2, di-heneicosanoyl phosphatidyl choline | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Scacioc, A, Wang, D, McKinley, G, Fernandez-Cid, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Burgess-Brown, N.A, van Putte, W, Duerr, K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
6SPB
 
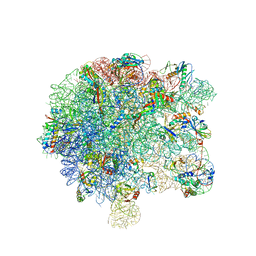 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate with a mutation in uL6 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PT4
 
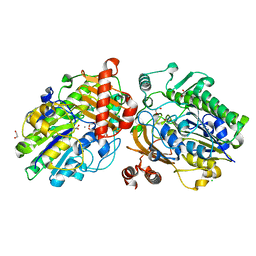 | | Crystal structure of apo PsS1_NC | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2019-07-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6MJ0
 
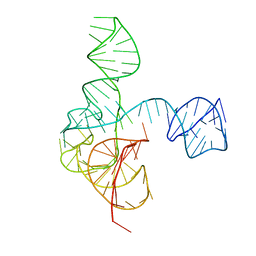 | | Crystal structure of the complete turnip yellow mosaic virus 3'UTR | | Descriptor: | RNA (101-MER) | | Authors: | Hartwick, E.W, Costantino, D.A, MacFadden, A, Nix, J.C, Tian, S, Das, R, Kieft, J.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ribosome-induced RNA conformational changes in a viral 3'-UTR sense and regulate translation levels.
Nat Commun, 9, 2018
|
|
5NMK
 
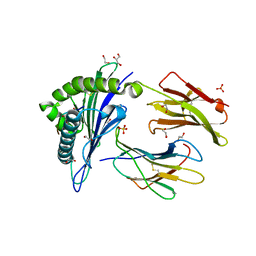 | | HLA A02 presenting SLFNTIAVL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2017-04-06 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Dual Molecular Mechanisms Govern Escape at Immunodominant HLA A2-Restricted HIV Epitope.
Front Immunol, 8, 2017
|
|
7AUO
 
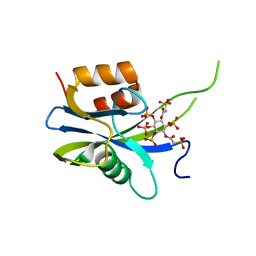 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PA-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(2-oxoethane-2,1-diyl)]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUT
 
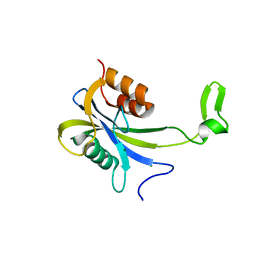 | |
