4R1Y
 
 | | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitor | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3-(diethylamino)propyl (3-{[5-(3,4-dimethoxyphenyl)-2-oxo-2H-1,3,4-thiadiazin-3(6H)-yl]methyl}phenyl)carbamate, Hepatocyte growth factor receptor | | Authors: | Blaukat, A, Bladt, F, Friese-Hamim, M, Knuehl, C, Fittschen, C, Graedler, U, Meyring, M, Dorsch, D, Stieber, F, Schadt, O. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6VIU
 
 | |
2I5B
 
 | | The crystal structure of an ADP complex of Bacillus subtilis pyridoxal kinase provides evidence for the parralel emergence of enzyme activity during evolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphomethylpyrimidine kinase | | Authors: | Newman, J.A, Das, S.K, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of an ADP Complex of Bacillus subtilis Pyridoxal Kinase Provides Evidence for the Parallel Emergence of Enzyme Activity During Evolution.
J.Mol.Biol., 363, 2006
|
|
5K7Y
 
 | | Crystal structure of enzyme in purine metabolism | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Skerlova, J, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2016-05-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interface modulation causes misregulation of purine 5 -nucleotidase in relapsed leukemia.
Bmc Biol., 14, 2016
|
|
2HY9
 
 | | Human telomere DNA quadruplex structure in K+ solution hybrid-1 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Punchihewa, C, Jones, R.A, Hurley, L, Yang, D. | | Deposit date: | 2006-08-04 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation.
Nucleic Acids Res., 35, 2007
|
|
2VGE
 
 | | Crystal structure of the C-terminal region of human iASPP | | Descriptor: | RELA-ASSOCIATED INHIBITOR | | Authors: | Robinson, R.A, Lu, X, Jones, E.Y, Siebold, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Studies of Aspp Proteins Reveal Differential Binding to P53, P63 and P73
Structure, 16, 2008
|
|
6PFL
 
 | | Crystal structure of Human HUWE1 WWE domain in complex with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, E3 ubiquitin-protein ligase HUWE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Dong, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Human HUWE1 WWE domain in complex with ADPR
to be published
|
|
2VH2
 
 | | Crystal structure of cell divison protein FtsQ from Yersinia enterecolitica | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Nanninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
6IVK
 
 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
4QQX
 
 | | Crystal structure of T. fusca Cas3-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6I4I
 
 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Mg-K-ADP-AlFn state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
5KBU
 
 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
3E5O
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4R7O
 
 | | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci
To be Published
|
|
7ZJ3
 
 | | Structure of TRIM2 RING domain in complex with UBE2D1~Ub conjugate | | Descriptor: | Polyubiquitin-C, Tripartite motif-containing protein 2, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Esposito, D, Garza-Garcia, A, Dudley-Fraser, J, Rittinger, K. | | Deposit date: | 2022-04-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity.
Nat Commun, 13, 2022
|
|
2I65
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
8QT7
 
 | |
3E77
 
 | | Human phosphoserine aminotransferase in complex with PLP | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase | | Authors: | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, S.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Wisniewska, M, Weigelt, J, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human phosphoserine aminotransferase in complex with PLP
TO BE PUBLISHED
|
|
4QX2
 
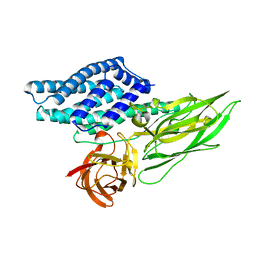 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3E8B
 
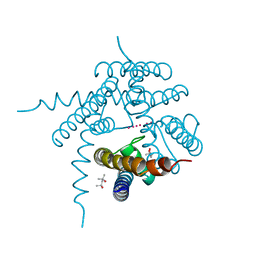 | |
5DGJ
 
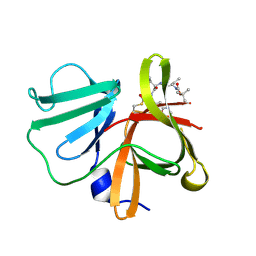 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
5JRU
 
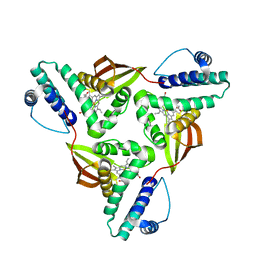 | | Crystal structure of Fe(II) unliganded H-NOX protein from C. subterraneus | | Descriptor: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J, Hespen, C, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX.
Acs Chem.Biol., 11, 2016
|
|
2I10
 
 | | Putative TetR transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | P-NITROPHENOL, Putative TetR transcriptional regulator, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-09-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TetR transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
