6PV8
 
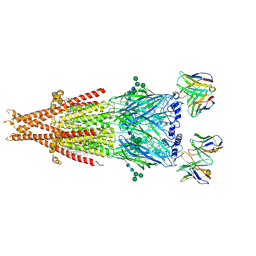 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with AT-1001 | | Descriptor: | (3-endo)-N-(2-bromophenyl)-9-methyl-9-azabicyclo[3.3.1]nonan-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
7BCR
 
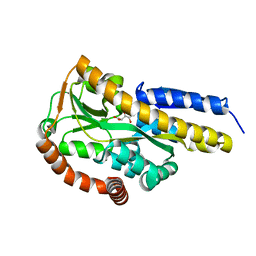 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with galactonate | | Descriptor: | L-galactonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
3GLY
 
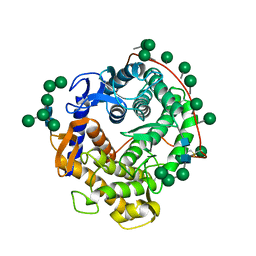 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-06-03 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
7VF1
 
 | |
8VBK
 
 | | Structure of bovine anti-HIV Fab ElsE5 | | Descriptor: | Bovine Fab ElsE5 heavy chain, Bovine Fab ElsE5 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultra-long CDRH3 repertoire
To Be Published
|
|
7BCO
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with D-foconate | | Descriptor: | (2R,3S,4S,5S)-2,3,4,5-tetrahydroxyhexanoic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
6SSK
 
 | |
5U82
 
 | | Crystal structure of a MerB-triethyltin complex | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5CE5
 
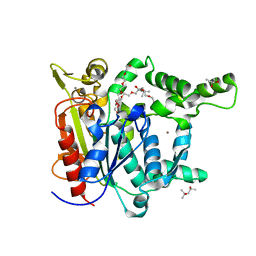 | | Probing the roles of two tryptophans surrounding the unique zinc coordination site in lipase family I.5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CALCIUM ION, ... | | Authors: | Sezerman, O.U, Podjarny, A.D, Emel, T, Cousido-Siah, A, Mitschler, A. | | Deposit date: | 2015-07-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the roles of two tryptophans surrounding the unique zinc coordination site in lipase family I.5.
Proteins, 84, 2016
|
|
6SZ2
 
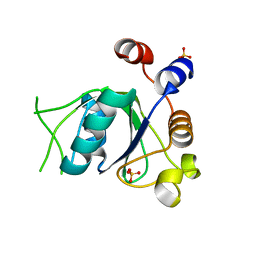 | | Crystal structure of YTHDC1 with fragment 3 (DHU_DC1_149) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methyl-1,6-naphthyridin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
7YOF
 
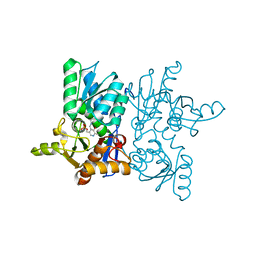 | |
3GM6
 
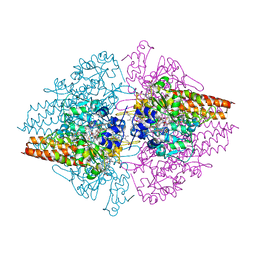 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with phosphate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of octaheme cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens in a complex with phosphate
Crystallography Reports, 55, 2010
|
|
7BCN
 
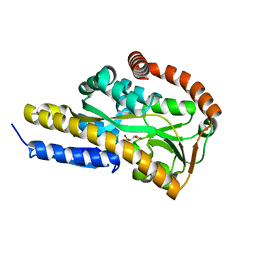 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with Xylonic acid | | Descriptor: | D-xylonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
5U8E
 
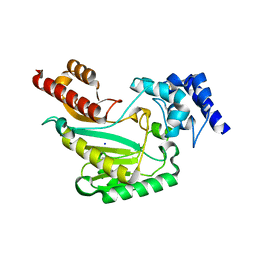 | | Crystal Structure of substrate-free arginine kinase from spider Polybetes pythagoricus | | Descriptor: | SODIUM ION, arginine kinase | | Authors: | Lopez-Zavala, A.A, Garcia, C.F, Hernadez-Paredes, J, Sotelo-Mundo, R.R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus.
PeerJ, 5, 2017
|
|
8VBJ
 
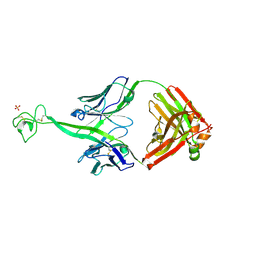 | | Structure of bovine anti-HIV Fab ElsE2 | | Descriptor: | Bovine Fab ElsE2 heavy chain, Bovine Fab ElsE2 light chain, POTASSIUM ION, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultra-long CDRH3 repertoire
To Be Published
|
|
5CFO
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'rotated' open conformation | | Descriptor: | Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
7BJ5
 
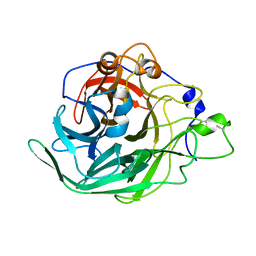 | | Inulosucrase from Halalkalicoccus jeotgali | | Descriptor: | Levansucrase | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
7VAX
 
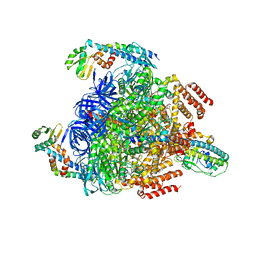 | | V1EG of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state1-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
4QJ3
 
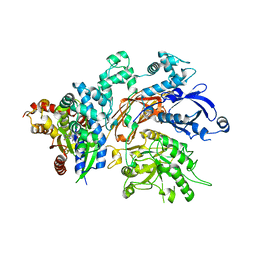 | | Structure of a fragment of human phospholipase C-beta3 delta472-559, in complex with Galphaq | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Structure, 22, 2014
|
|
5MNL
 
 | |
6SW1
 
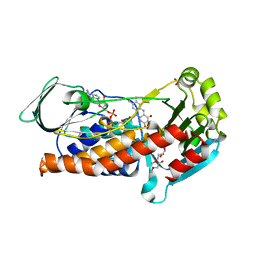 | | Crystal Structure of P. aeruginosa PqsL: R41Y, I43R, G45R, C105G mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mattevi, A, Rovida, S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoinduced monooxygenation involving NAD(P)H-FAD sequential single-electron transfer.
Nat Commun, 11, 2020
|
|
7KRU
 
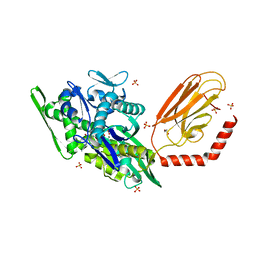 | |
4YSX
 
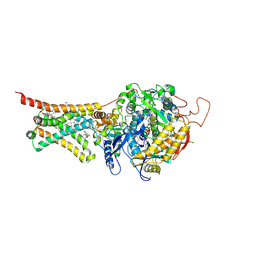 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with the specific inhibitor NN23 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
7YOL
 
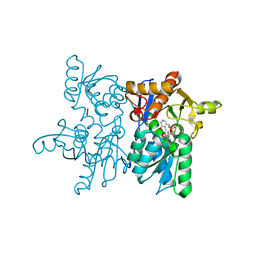 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
8RM6
 
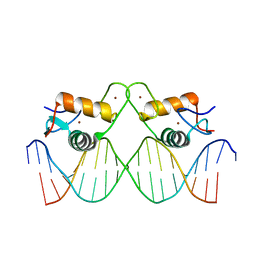 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: C3(1)ARE | | Descriptor: | C3(1)ARE_Chain C, C3(1)ARE_Chain D, Isoform 2 of Androgen receptor, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
