7R1C
 
 | |
6P6S
 
 | | HCV NS3/4A protease domain of genotype 3a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
7R4I
 
 | | The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 2.15, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
4EU6
 
 | | Succinyl-CoA:acetate CoA-transferase (AarCH6) in complex with CoA, acetate, and covalent acetylglutamyl anhydride and glutamyl-CoA thioester adducts | | Descriptor: | ACETATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Mullins, E.A, Kappock, T.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-10 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Crystal Structures of Acetobacter aceti Succinyl-Coenzyme A (CoA):Acetate CoA-Transferase Reveal Specificity Determinants and Illustrate the Mechanism Used by Class I CoA-Transferases.
Biochemistry, 51, 2012
|
|
6XNM
 
 | |
5B6V
 
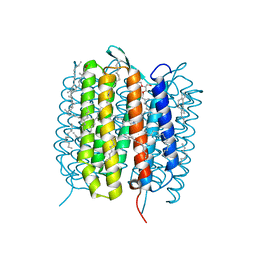 | | A three dimensional movie of structural changes in bacteriorhodopsin: resting state structure | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nango, E, Royant, A, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6ERK
 
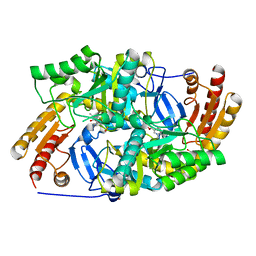 | | Crystal structure of diaminopelargonic acid aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Stekhanova, T.N, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2017-10-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diaminopelargonic acid transaminase from Psychrobacter cryohalolentis is active towards (S)-(-)-1-phenylethylamine, aldehydes and alpha-diketones.
Appl. Microbiol. Biotechnol., 102, 2018
|
|
1AI8
 
 | | HUMAN ALPHA-THROMBIN TERNARY COMPLEX WITH THE EXOSITE INHIBITOR HIRUGEN AND ACTIVE SITE INHIBITOR PHCH2OCO-D-DPA-PRO-BOROMPG | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN IIIB, ... | | Authors: | Skordalakes, E, Dodson, G, Elgendy, S, Goodwin, C.A, Green, D, Tyrrel, R, Scully, M.F, Freyssinet, J, Kakkar, V.V, Deadman, J. | | Deposit date: | 1997-05-01 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships.
Protein Sci., 1, 1992
|
|
5FLP
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 5-[(2-chloranylphenoxy)methyl]-1H-1,2,3,4-tetrazole, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5J6S
 
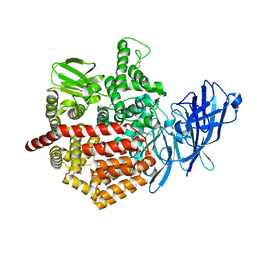 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | Descriptor: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | Deposit date: | 2016-04-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
6P6O
 
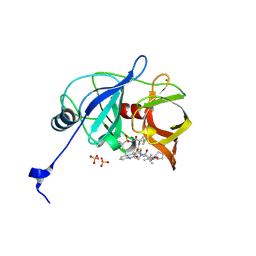 | | HCV NS3/4A protease domain of genotype 1a D168E in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, CHLORIDE ION, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | HCV NS3/4A protease domain of genotype 1a in complex with glecaprevir
To Be Published
|
|
3LAH
 
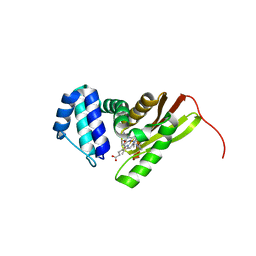 | | Structural insights into the molecular mechanism of H-NOX activation | | Descriptor: | IMIDAZOLE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Olea Jr, C, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the molecular mechanism of H-NOX activation.
Protein Sci., 19, 2010
|
|
7XX6
 
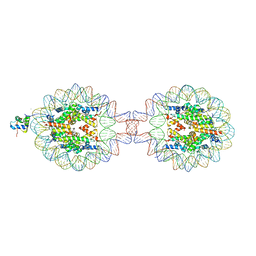 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
6RVE
 
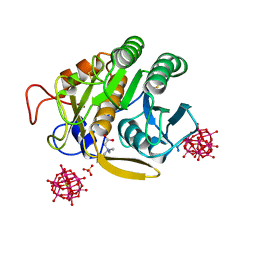 | | Co-substituted beta-Keggin bound to Proteinase K solved by MR | | Descriptor: | Co-substituted beta-Keggin, Proteinase K, SULFATE ION, ... | | Authors: | Breibeck, J, Bijelic, A, Rompel, A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transition metal-substituted Keggin polyoxotungstates enabling covalent attachment to proteinase K upon co-crystallization.
Chem.Commun.(Camb.), 55, 2019
|
|
5FQF
 
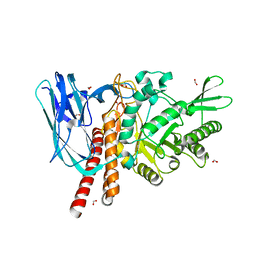 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
4XS6
 
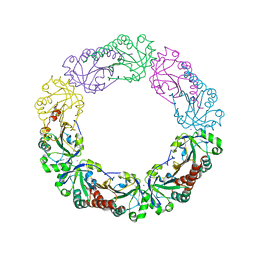 | | Salmonella typhimurium AhpC W81F mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, POTASSIUM ION | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
8GFO
 
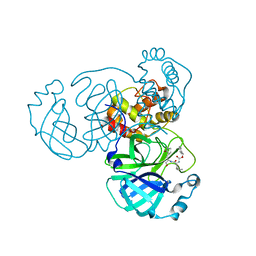 | |
4XSN
 
 | | Copper(II) bound to the Z-DNA form of d(CGCGCG) | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(BGM)P*CP*GP*CP*GP)-3') | | Authors: | Rohner, M, Medina-Molner, A, Spingler, B. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | N,N,O and N,O,N Meridional cis Coordination of Two Guanines to Copper(II) by d(CGCGCG)2.
Inorg.Chem., 55, 2016
|
|
8GFU
 
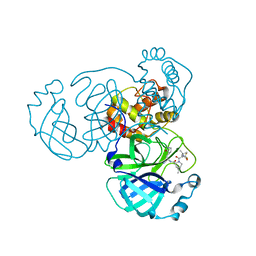 | |
3LCZ
 
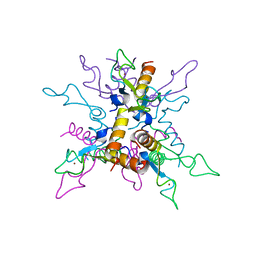 | | B.licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
8ASE
 
 | | Crystal structure of Thrombin in complex with macrocycle T3 | | Descriptor: | (8~{S},14~{S},18~{E})-8-[(4-chlorophenyl)methyl]-3,21-dithia-7,10,16-triazatricyclo[21.2.2.1^{10,14}]octacosa-1(26),18,23(27),24-tetraene-6,9,15-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chinellato, M, Angelini, A, Nielsen, A, Heinis, C, Cendron, L. | | Deposit date: | 2022-08-19 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thrombin in complex with optimized macrocycles T1 and T3
To Be Published
|
|
5FV1
 
 | | Crystal structure of hVEGF in complex with VK domain antibody | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR A, VK DOMAIN ANTIBODY | | Authors: | Chung, C, Walker, A. | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel Interaction Mechanism of a Domain Antibody Based Inhibitor of Human Vascular Endothelial Growth Factor with Greater Potency Than Ranibizumab and Bevacizumab and Improved Capacity Over Aflibercept.
J.Biol.Chem., 291, 2016
|
|
7RCC
 
 | |
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5FLS
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | (E)-3-(4-chlorophenyl)but-2-enoic acid, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
