5MUW
 
 | | Atomic structure of P4 packaging enzyme fitted into a localized reconstruction of bacteriophage phi6 vertex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
8DQH
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 24 hours of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
2ELQ
 
 | | Solution structure of the 14th C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 14th C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
1UZC
 
 | | THE STRUCTURE OF AN FF DOMAIN FROM HUMAN HYPA/FBP11 | | Descriptor: | HYPOTHETICAL PROTEIN FLJ21157 | | Authors: | Allen, M.D, Jemth, P, Friedler, A, Schon, O, Bycroft, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of an Ff Domain from Human Hypa/Fbp11.
J.Mol.Biol., 323, 2002
|
|
8E5W
 
 | | Crystal structure of dehydroalanine Hip1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PALMITIC ACID, Protease, ... | | Authors: | Goldfarb, N.E, Brooks, C.L, Ostrov, D.A. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 angstrom crystal structure of the Mycobacterium tuberculosis serine hydrolase, Hip1, in its anhydro-form (Anhydrohip1).
Biochem.Biophys.Res.Commun., 630, 2022
|
|
5MV5
 
 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DQJ
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (AST) bound to ATP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8U66
 
 | | Firmicutes Rubisco | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Rubisco | | Authors: | Kaeser, B.P, Liu, A.K, Shih, P.M. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Deep-branching evolutionary intermediates reveal structural origins of form I rubisco.
Curr.Biol., 33, 2023
|
|
6OPT
 
 | | HIV-1 Protease NL4-3 V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
7ALH
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
6GP5
 
 | | Beta-1,4-galactanase from Bacteroides thetaiotaomicron | | Descriptor: | Arabinogalactan endo-beta-1,4-galactanase | | Authors: | Hekelaar, J, Boger, M, Leeuwen van, S.S, Lammerts van Bueren, A, Dijkhuizen, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and functional characterization of a family GH53 beta-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.
J. Struct. Biol., 205, 2019
|
|
8E3V
 
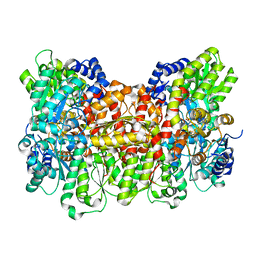 | |
4B2U
 
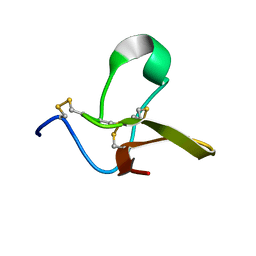 | | S67, A spider venom toxin peptide from Sicarius dolichocephalus | | Descriptor: | S67 | | Authors: | Loening, N.M, Wilson, Z.N, Zobel-Thropp, P.A, Binford, G.J. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Two Homologous Venom Peptides from Sicarius Dolichocephalus.
Plos One, 8, 2013
|
|
1YDM
 
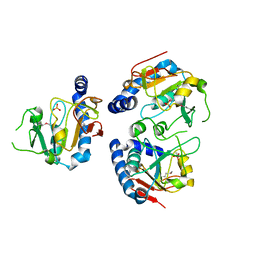 | | X-Ray structure of Northeast Structural Genomics target SR44 | | Descriptor: | Hypothetical protein yqgN, SULFATE ION | | Authors: | Kuzin, A.P, Jianwei, S, Vorobiev, S, Acton, T, Xia, R, Ma, L.-C, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of Northeast Structural Genomics target SR44
To be Published
|
|
6H2E
 
 | |
8QG0
 
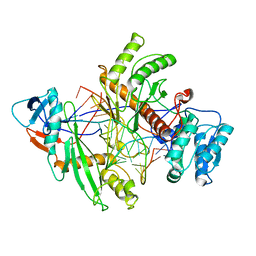 | | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 17 nt RNA guide and 17 nt DNA target | | Descriptor: | AfAgo-N protein, DNA target 17 nt, Piwi protein, ... | | Authors: | Manakova, E.N, Zaremba, M, Pocevicuite, R, Golovinas, E, Zagorskaite, E, Silanskas, A. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
4BKT
 
 | | von Hippel Lindau protein:ElonginB:ElonginC complex, in complex with (2S,4R)-N-methyl-1-[2-(3-methyl-1,2-oxazol-5-yl)ethanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide | | Descriptor: | (2S,4R)-N-methyl-1-[2-(3-methyl-1,2-oxazol-5-yl)ethanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, ARGININE, GLUTAMIC ACID, ... | | Authors: | Van Molle, I, Dias, D.M, Baud, M, Galdeano, C, Geraldes, C.F.G.C, Ciulli, A. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Is NMR Fragment Screening Fine-Tuned to Assess Druggability of Protein-Protein Interactions?
Acs Med.Chem.Lett., 5, 2014
|
|
6YAN
 
 | | Mammalian 48S late-stage translation initiation complex with histone 4 mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
8Q34
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the ligand ZZ001229a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(1~{H}-imidazo[4,5-b]pyridin-2-ylmethyl)-3-(3-methyl-1,2-diazirin-3-yl)propanamide | | Authors: | MacLean, E.M, Gao, Q, Williams, E, Balcomb, B.H, von Delft, F, Bajusz, D, Keeley, A, Abranyi-Balogh, P, Koekemoer, L, Keseru, G.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein binding sites by photoreactive fragment pharmacophores.
Commun Chem, 7, 2024
|
|
5MDN
 
 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6EXD
 
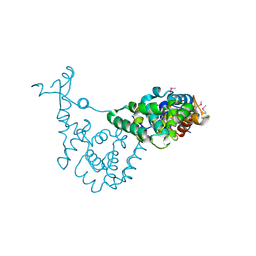 | |
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | Descriptor: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
6GS1
 
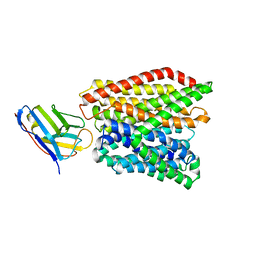 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
5J73
 
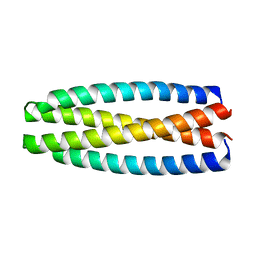 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | protein design 2L4HC2_9 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
8JQ5
 
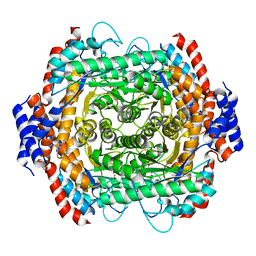 | |
