6VCG
 
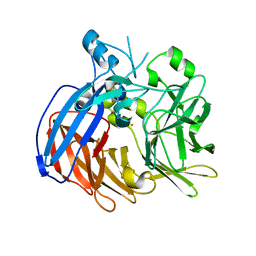 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | Descriptor: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
1K5A
 
 | | Crystal structure of human angiogenin double variant I119A/F120A | | Descriptor: | Angiogenin | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
5O1P
 
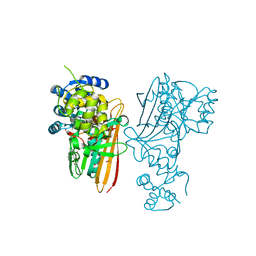 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
7M9Z
 
 | | HIV-1 Protease (I84V) in Complex with TMC-126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with TMC-126
To Be Published
|
|
6G2D
 
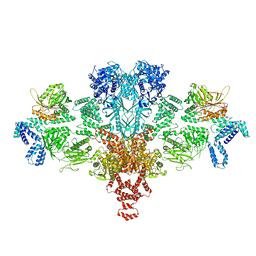 | | Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6UPT
 
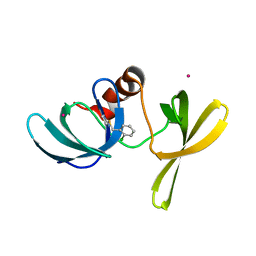 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706 | | Descriptor: | 2-((2-chlorobenzyl)thio)-4,5-dihydro-1H-imidazole, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706
to be published
|
|
2HFB
 
 | |
8CCN
 
 | | Filamentous actin II from Plasmodium falciparum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-2, MAGNESIUM ION | | Authors: | Kursula, I, Lopez, A.J. | | Deposit date: | 2023-01-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of Plasmodium actin II in the parasite mosquito stages.
Plos Pathog., 19, 2023
|
|
7SAH
 
 | |
8CAG
 
 | | Hypoxanthine-guanine phosphoribosyltransferase from E. coli | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Kostromina, M.A, Zayats, E.A, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hypoxanthine-guanine phosphoribosyltransferase from E. coli
To Be Published
|
|
9BDD
 
 | | Cryo-EM Structure of Non-Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
3I5I
 
 | | The crystal structure of squid myosin S1 in the presence of SO4 2- | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
7KVE
 
 | |
8OQW
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK | | Descriptor: | ATPase, GLYCEROL, SULFATE ION | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
3QRU
 
 | | CDK2 in complex with inhibitor NSK-MC1-12 | | Descriptor: | (5S)-N-methyl-5-(2-methylbutan-2-yl)-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, 1,2-ETHANEDIOL, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
6RX4
 
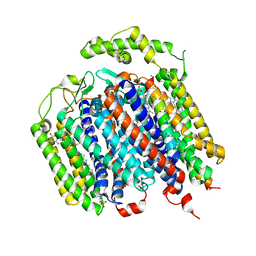 | | THE STRUCTURE OF BD OXIDASE FROM ESCHERICHIA COLI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Rasmussen, T, Boettcher, B, Thesseling, A, Friedrich, T. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Homologous bd oxidases share the same architecture but differ in mechanism.
Nat Commun, 10, 2019
|
|
6EPI
 
 | |
8OOC
 
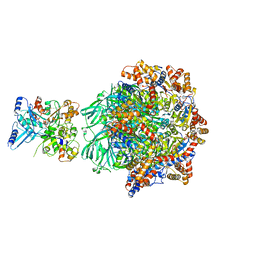 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8BZ3
 
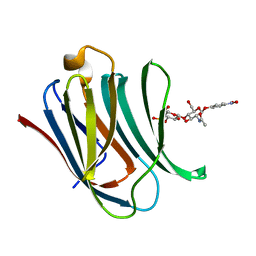 | | Structure od the carbohydrate reconition domain of Gal3 in comples with SAF-2-010 | | Descriptor: | Galectin-3, [(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{S})-5-acetamido-2-(hydroxymethyl)-6-(4-nitrophenoxy)-4-oxidanyl-oxan-3-yl]oxy-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-4-yl] hydrogen sulfate | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-12-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Selectively Modified Lactose and N -Acetyllactosamine Analogs at Three Key Positions to Afford Effective Galectin-3 Ligands.
Int J Mol Sci, 24, 2023
|
|
8OW7
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 in complex with N-acetylmuramic acid (MurNAc) | | Descriptor: | N-acetyl-beta-muramic acid, N-acetylglucosamine kinase, SULFATE ION | | Authors: | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
6UU6
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a UTP ("Old" crystal soaked with UTP, ddCTP, and dinucleotide ApG for 30 minutes) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.201 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
8OW9
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK in complex with N-acetylmuramic acid (MurNAc) | | Descriptor: | N-acetyl-beta-muramic acid, Putative novel MurNAc kinase | | Authors: | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8CCO
 
 | |
5NAZ
 
 | |
