5OAZ
 
 | |
2VGL
 
 | | AP2 CLATHRIN ADAPTOR CORE | | Descriptor: | ADAPTOR PROTEIN COMPLEX AP-2, ALPHA 2 SUBUNIT, AP-2 COMPLEX SUBUNIT BETA-1, ... | | Authors: | Owen, D.J, Collins, B.M, McCoy, A.J, Evans, P.R. | | Deposit date: | 2007-11-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture and Functional Model of the Endocytic Ap2 Complex
Cell(Cambridge,Mass.), 109, 2002
|
|
8BO4
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 1 | | Descriptor: | 4-(aminomethyl)-~{N}-[(2~{S})-1-oxidanylidene-3-phenyl-1-[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]amino]propan-2-yl]cyclohexane-1-carboxamide, Coagulation factor XIa light chain, GLYCEROL | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, M, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
8BO3
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR Asundexian | | Descriptor: | 4-[[(2~{S})-2-[4-[5-chloranyl-2-[4-(trifluoromethyl)-1,2,3-triazol-1-yl]phenyl]-5-methoxy-2-oxidanylidene-pyridin-1-yl]butanoyl]amino]-2-fluoranyl-benzamide, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
7SEC
 
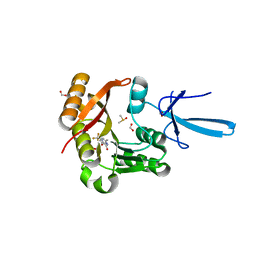 | | Crystal structure of human Fibrillarin in complex with compound 1a | | Descriptor: | 2-[(8S)-4-oxo-2-(trifluoromethyl)-4,5-dihydropyrazolo[1,5-a]pyrazin-6-yl]acetamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
8BO5
 
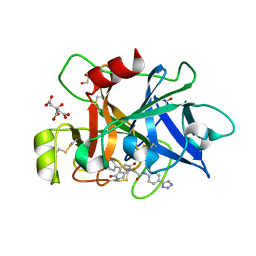 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 3 | | Descriptor: | 2-[4-(3-chlorophenyl)-2,5-bis(oxidanylidene)pyrrolo[3,4-b]pyridin-1-yl]-~{N}-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]ethanamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
8BO6
 
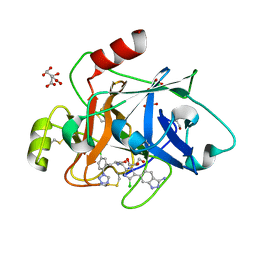 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 2 | | Descriptor: | (~{E})-~{N}-[[5-(3-azanyl-1~{H}-indazol-6-yl)-4-chloranyl-1~{H}-imidazol-2-yl]methyl]-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
7NUI
 
 | |
8BO7
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 34 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[[(2~{S})-2-[4-(5-chloranyl-2-cyano-phenyl)-3-methoxy-6-oxidanylidene-2,5-dihydropyridin-1-yl]-3-[(2~{S})-oxan-2-yl]propanoyl]amino]benzoic acid, CITRIC ACID, ... | | Authors: | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
6PFV
 
 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6RWK
 
 | |
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
8TCO
 
 | | HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CS2it1p2_F7K Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2023-07-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Single-cell analysis of memory B cells from top neutralizers reveals multiple sites of vulnerability within HCMV Trimer and Pentamer.
Immunity, 56, 2023
|
|
8T9R
 
 | |
7SE8
 
 | | Crystal structure of human Fibrillarin in complex with fragment 1 from cocktail soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
5NSW
 
 | | Xenon for tunnelling analysis of the efflux pump component OprN. | | Descriptor: | Multidrug efflux outer membrane protein OprN, NICKEL (II) ION, PALMITIC ACID, ... | | Authors: | Phan, G, Prange, T, Enguene Ntsogo, Y.V, Garnier, C, Ducruix, A, Broutin, I. | | Deposit date: | 2017-04-27 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
7SE9
 
 | | Crystal structure of human Fibrillarin in complex with compound 1 from single soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
6Y9C
 
 | | The structure of a quaternary ammonium Rieske monooxygenase reveals insights into carnitine oxidation by gut microbiota and inter-subunit electron transfer | | Descriptor: | CARNITINE, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterisation of an unusual cysteine pair in the Rieske carnitine monooxygenase CntA catalytic site.
Febs J., 2023
|
|
8TEA
 
 | |
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
5WEC
 
 | |
6PPS
 
 | | A blue light illuminated LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Fernandez, I, Shin, H, Gunawardana, S, Otero, L.H, Cerutti, M.L, Yang, X, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
7AH3
 
 | | Kinase domain of cSrc in complex with a pyrazolopyrimidine | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Dello Iacono, L, Kleinboelting, S, Fallacara, A.L, Rauh, D. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the binding of pyrazolopyrimidines to Src kinase
To Be Published
|
|
4A8I
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
