7GJ1
 
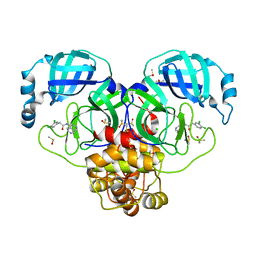 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with VLA-UNK-82501c2c-1 (Mpro-P0153) | | Descriptor: | 2-(3,4-dichlorophenyl)-N-(2,7-naphthyridin-4-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8FQC
 
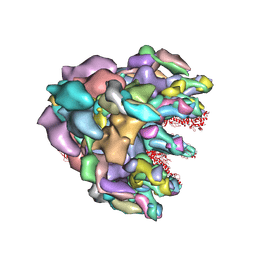 | | Structure of baseplate with receptor binding complex of Agrobacterium phage Milano | | Descriptor: | Baseplate Centerpiece, gp25, Baseplate Central Spike, ... | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
8AWU
 
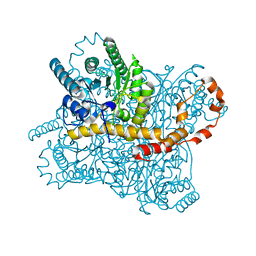 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 250ms | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
7GJH
 
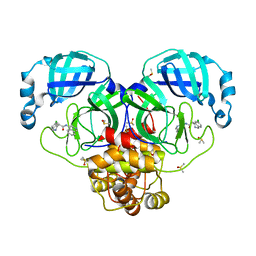 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-f2e727cd-5 (Mpro-P0240) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8AWF
 
 | | Xylose Isomerase in 80% relative humidity environment | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-08-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
7GJX
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-3ccb8ef6-1 (Mpro-P0744) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4QKX
 
 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4X9S
 
 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
8FOU
 
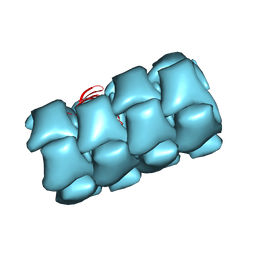 | | Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-tube | | Descriptor: | Virion-associated protein | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
7Q9B
 
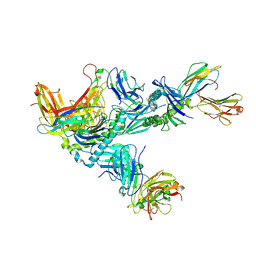 | | MHC Class I A02 Allele presenting EAAGIGILTV, in complex with Mel8 TCR | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLU-ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7GKE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-dd3ad2b5-3 (Mpro-P0851) | | Descriptor: | (4S)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroisoquinoline-2,4(1H)-dicarboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7Q99
 
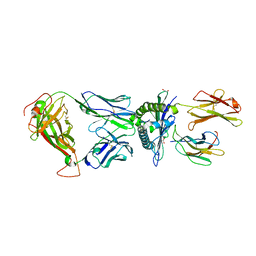 | | MHC Class I A02 Allele presenting NLSALGIFST, in complex with Mel5 TCR | | Descriptor: | 1,2-ETHANEDIOL, ASN-LEU-SER-ALA-LEU-GLY-ILE-PHE-SER-THR, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
3L7J
 
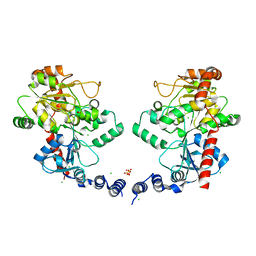 | |
6XYZ
 
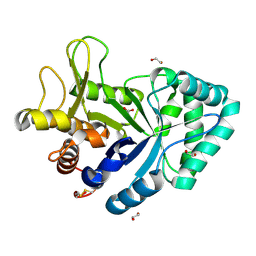 | | Crystal structure of the GH18 chitinase ChiB from the chitin utilization locus of Flavobacterium johnsoniae | | Descriptor: | 1,2-ETHANEDIOL, Candidate chitinase Glycoside hydrolase family 18, FORMIC ACID | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V, Pope, P, Branden, G, Larsbrink, J. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
6PKY
 
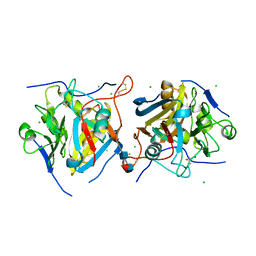 | | Guinea pig N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) catalytic domain auto-inhibited by pro-peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2019-06-30 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
8FOY
 
 | | Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-sheath | | Descriptor: | Tail sheath protein | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
7GKU
 
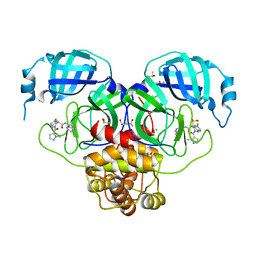 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-4223bc15-40 (Mpro-P1079) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(1-methyl-1H-pyrazole-5-carbonyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4XDT
 
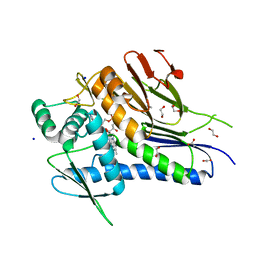 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, a bifunctional FMN transferase/FAD pyrophosphatase, N55Y mutant, FAD bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
7Q98
 
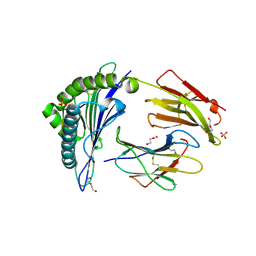 | | MHC Class I A02 Allele presenting NLSALGIFST | | Descriptor: | 1,2-ETHANEDIOL, ASN-LEU-SER-ALA-LEU-GLY-ILE-PHE-SER-THR, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7Q9A
 
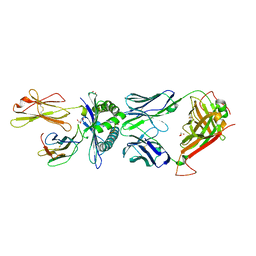 | | MHC Class I A02 Allele presenting LLLGIGILVL, in complex with Mel5 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Human Mel5 T Cell Receptor, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7GLA
 
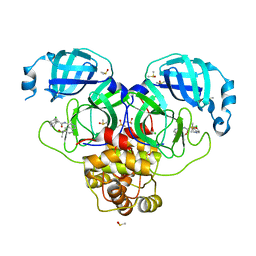 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-86c60949-2 (Mpro-P1812) | | Descriptor: | (4R)-6-chloro-N-[6-(2-hydroxypropan-2-yl)isoquinolin-4-yl]-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7QOF
 
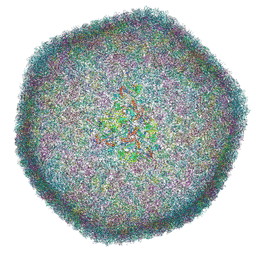 | | Icosahedral capsid of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Head fiber dimer protein gp29, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7GLQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-8df914d1-2 (Mpro-P2007) | | Descriptor: | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4QJ9
 
 | |
7QOH
 
 | | Unique vertex of the phicrAss001 virion with C5 symmetry imposed | | Descriptor: | Auxiliary capsid protein gp36, Head fiber trimer protein gp21, MAGNESIUM ION, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
