6OSU
 
 | | Crystal Structure of the D-alanyl-D-alanine carboxypeptidase DacD from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase (Penicillin binding protein) family protein | | Authors: | Kim, Y, Stogios, P, Skarina, T, Di, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the D-alanyl-D-alanine carboxypeptidase DacD from Francisella tularensis
To Be Published
|
|
6TXE
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dTMPNPP and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
6XRX
 
 | | Crystal structure of the mosquito protein AZ1 as an MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Foo, A.C.Y. | | Deposit date: | 2020-07-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mosquito protein AEG12 displays both cytolytic and antiviral properties via a common lipid transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6OUI
 
 | |
5M1D
 
 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
8F0A
 
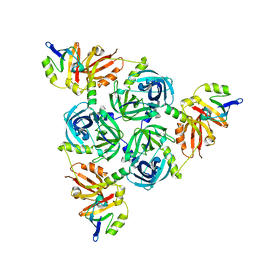 | | Client-bound structure of a DegP trimer within a 12mer cage | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
6XIY
 
 | | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to methyl 2-(acetylamino)-2-deoxy-1-thio-alpha-D-galactopyranose | | Descriptor: | C-type lectin domain family 10 member A, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Birrane, G, Murphy, P.V, Gabba, A, Luz, J.G. | | Deposit date: | 2020-06-22 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc and the Tumor-Associated Tn Antigen.
Biochemistry, 60, 2021
|
|
6X6F
 
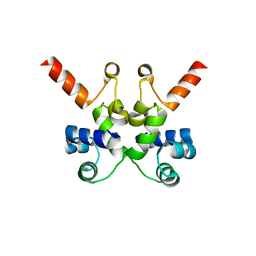 | | The structure of Pf6r from the filamentous phage Pf6 of Pseudomonas aeruginosa PA01 | | Descriptor: | NITRATE ION, Pf6r | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6UQV
 
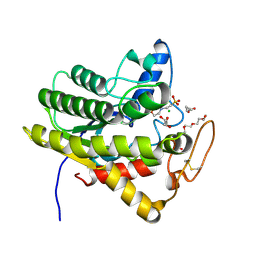 | | Crystal structure of ChoE, a bacterial acetylcholinesterase from Pseudomonas aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BUTANOIC ACID, CHLORIDE ION, ... | | Authors: | Shi, R, Pham, V.D, To, T.A. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
8ENI
 
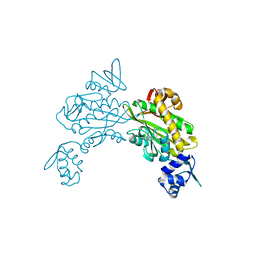 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Wilce, M.C.J, Cini, D.A. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6OE7
 
 | | Crystal structure of HMCES cross-linked to DNA abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*(DRZ)P*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6XDB
 
 | | Crystal structure of IRE1a in complex with G-6904 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-N-(6-chloro-2-fluoro-3-{[(2-fluorophenyl)sulfonyl]amino}phenyl)-6-(1,3-dimethyl-1H-pyrazol-4-yl)quinazoline-8-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.A, Weiru, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
8F43
 
 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
5LI9
 
 | | Structure of a nucleotide-bound form of PKCiota core kinase domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
7AG8
 
 | |
7AHB
 
 | | Acyltransferase domain of the polyketide synthase PpsC of Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Phthiocerol synthesis polyketide synthase type I PpsC, SODIUM ION, ... | | Authors: | Faille, A, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
6OFW
 
 | | Dihydropteroate synthase from Xanthomonas albilineans in complex with 7,8-dihydropteroate | | Descriptor: | 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}benzoic acid, Dihydropteroate synthase, SULFATE ION | | Authors: | Oliveira, A.A, Guido, R.V.C, Maluf, F.V, Bueno, R.V, Lima, G.M.A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dihydropteroate synthase from Xanthomonas albilineans in complex with 7,8-dihydropteroate
To Be Published
|
|
7NLH
 
 | |
8SKB
 
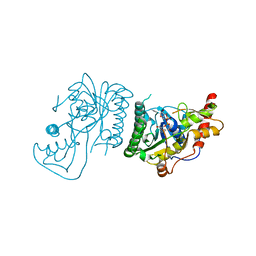 | | Crystal Structure of GDP-mannose 3,5 epimerase de Myrciaria dubia in complex with NAD | | Descriptor: | GDP-mannose 3,5 epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
6XM9
 
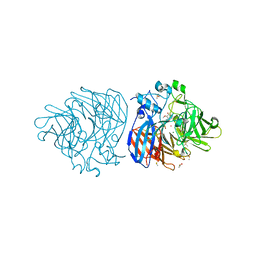 | | Crystal structure of vanillin bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETATE ION, COBALT (II) ION, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XM7
 
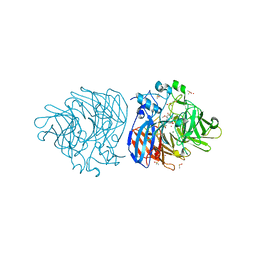 | | Crystal structure of DCA-S bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | (2E)-3-{4-hydroxy-3-[(E)-2-(4-hydroxy-3-methoxyphenyl)ethenyl]-5-methoxyphenyl}prop-2-enoic acid, COBALT (II) ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
7PY7
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
6VA3
 
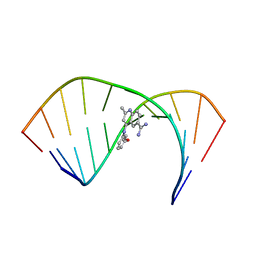 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MQC | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
7PYK
 
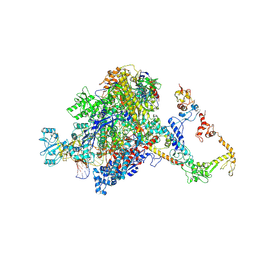 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
