6MJ6
 
 | | Crystal structure of the mCD1d/xxx (JJ166) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
7EHS
 
 | | Levansucrase from Brenneria sp. EniD 312 | | Descriptor: | GLYCEROL, Levansucrase, NONAETHYLENE GLYCOL, ... | | Authors: | Xu, W, Ni, D.W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
8UP8
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F, complex with L-Asp) | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
5JGW
 
 | | Crystal structure of maize AKR4C13 in complex with NADP and acetate | | Descriptor: | ACETATE ION, Aldose reductase, AKR4C13, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of maize AKR4C13 in complex with NADP and acetate
To Be Published
|
|
8B90
 
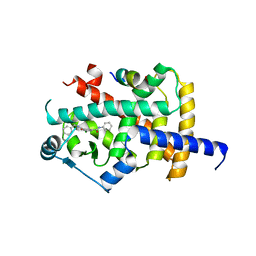 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 7d) | | Descriptor: | 5-chloranyl-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
5NAY
 
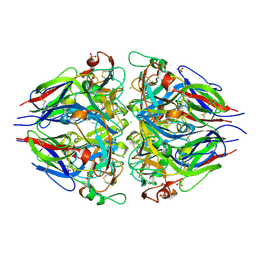 | |
6F9J
 
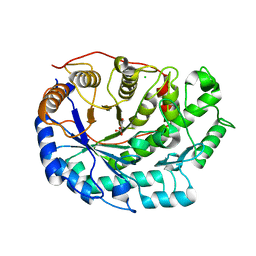 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-mannopyranose-(1-4)-1-DEOXYNOJIRIMYCIN | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
6X91
 
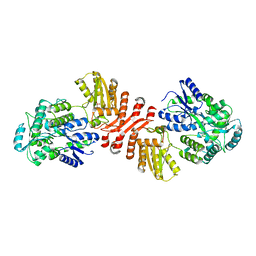 | | Crystal structure of MBP-fused human APOBEC1 | | Descriptor: | CACODYLATE ION, Maltodextrin-binding protein, C->U-editing enzyme APOBEC-1 chimera, ... | | Authors: | Wolfe, A.D, Li, S.-X, Chen, X.S. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity.
NAR Cancer, 2, 2020
|
|
9BKJ
 
 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | Descriptor: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
6GU2
 
 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6FAX
 
 | | Complex of Human CD40 Ectodomain with Lob 7.4 Fab | | Descriptor: | Lob 7.4 heavy chain, Lob 7.4 light chain, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Orr, C.M, Tews, I, Pearson, A.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Complex Interplay between Epitope Specificity and Isotype Dictates the Biological Activity of Anti-human CD40 Antibodies.
Cancer Cell, 33, 2018
|
|
8JVF
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8GZ5
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
6P0E
 
 | | Human DNA Ligase 1 (E346A,E592A) bound to adenylated DNA containing an 8-oxo guanine:adenine base-pair | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
8UOW
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
5JJ5
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to hydroxymate siderophore ferrioxamine E and iron(III) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, ABC transporter substrate-binding protein-iron transport, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kurdritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to hydroxymate siderophore ferrioxamine E and iron(III)
To Be Published
|
|
6ZN0
 
 | | Crystal structure of cAMP-dependent protein kinase A (CHO PKA) in complex with isonicotinamidine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIMETHYL SULFOXIDE, ISONICOTINAMIDINE, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-07-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5N52
 
 | |
7U0C
 
 | | Crystal structure of broadly neutralizing antibody HEPC3.4 | | Descriptor: | HEPC3.4 Fab Heavy Chain, HEPC3.4 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Computational identification of HCV neutralizing antibodies with a common HCDR3 disulfide bond motif in the antibody repertoires of infected individuals.
Nat Commun, 13, 2022
|
|
5AIF
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample-Native | | Descriptor: | IMIDAZOLE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5JK0
 
 | |
6SLS
 
 | |
8UP3
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
7N2Z
 
 | | Crystal Structure of a de Novo Three-stranded Coiled Coil Peptide Containing Trigonal Pyrmidal Pb(II) complexes in the dual Tris-thiolate Site | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)2-(GRAND CoilSerL16CL23C)3, ... | | Authors: | Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Open Reading Frame 1 Protein of the Human Long Interspersed Nuclear Element 1 Retrotransposon Binds Multiple Equivalents of Lead.
J.Am.Chem.Soc., 143, 2021
|
|
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
