7U0Q
 
 | |
5L7G
 
 | | MCR IN COMPLEX WITH ligand | | Descriptor: | 1,2-ETHANEDIOL, Mineralocorticoid receptor, NCOA1 peptide, ... | | Authors: | Edman, K, Aagaard, A, Backstrom, S, Xue, Y. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Drug Design of Mineralocorticoid Receptor Antagonists to Explore Oxosteroid Receptor Selectivity.
ChemMedChem, 12, 2017
|
|
7U0X
 
 | |
5NI8
 
 | | Ligand complex of RORg LBD | | Descriptor: | 2-(4-ethylsulfonylphenyl)-~{N}-[4-(2-phenylmethoxypyridin-3-yl)thiophen-2-yl]ethanamide, Nuclear receptor ROR-gamma, SODIUM ION, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2017-03-23 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
7U4C
 
 | | Borrelia burgdorferi HtpG N-terminal domain (1-228) in complex with BX-2819 | | Descriptor: | Chaperone protein HtpG, ethyl (4-{3-[2,4-dihydroxy-5-(1-methylethyl)phenyl]-5-sulfanyl-4H-1,2,4-triazol-4-yl}benzyl)carbamate | | Authors: | Kowalewski, M.E, Lietzan, A, Haystead, T, Redinbo, M.R. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting Borrelia burgdorferi HtpG with a berserker molecule, a strategy for anti-microbial development.
Cell Chem Biol, 2023
|
|
8JB8
 
 | | Crystal structure of CMY-185 complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
7U4P
 
 | |
5LAU
 
 | | Oceanobacillus iheyensis macrodomain mutant G37V with ADPR | | Descriptor: | GLYCEROL, MacroD-type macrodomain, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5LAZ
 
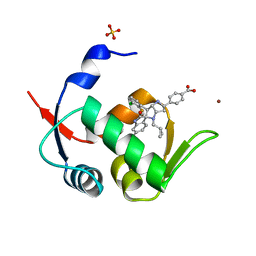 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND BI-0252 | | Descriptor: | 4-[(2~{R},3~{a}~{S},5~{S},6~{S},6~{a}~{S})-6'-chloranyl-6-(3-chloranyl-2-fluoranyl-phenyl)-4-(cyclopropylmethyl)-2'-oxidanylidene-spiro[1,2,3,3~{a},6,6~{a}-hexahydropyrrolo[3,2-b]pyrrole-5,3'-1~{H}-indole]-2-yl]benzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
6RU4
 
 | |
5KO3
 
 | |
6GA4
 
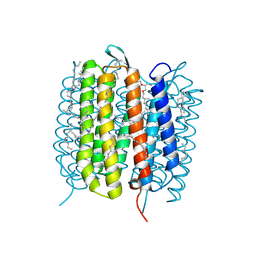 | | Bacteriorhodopsin, 1 ps state, real-space refined against 15% extrapolated map | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAD
 
 | | BACTERIORHODOPSIN, 530 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6RX7
 
 | |
5KRO
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Methyl(phenyl)amino-substituted Estrogen, (8R,9S,13S,14S,17S)-13-methyl-17-(methyl(phenyl)amino)-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (8~{R},9~{S},13~{S},14~{S},17~{S})-13-methyl-17-[methyl(phenyl)amino]-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthren-3-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6GOB
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, Lysozyme C, N,N-pyridylbenzimidazole derivative-Pd complex, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
4ZFY
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with ALPHA-METHYL-D-GALACTOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, methyl alpha-D-galactopyranoside, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
6GUY
 
 | | Room temperature structure of Archaerhodopsin-3 via LCP extruder using synchrotron radiation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Axford, D, Bada Juarez, J.F, Vinals, J, Watts, A. | | Deposit date: | 2018-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Room temperature structure of Archaerhodopsin-3 via LCP extruder using synchrotron radiation
To Be Published
|
|
5GAH
 
 | | RNC in complex with SRP with detached NG domain | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
6TP6
 
 | | Crystal structure of the Orexin-1 receptor in complex with filorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CHLORIDE ION, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5L31
 
 | |
5KJM
 
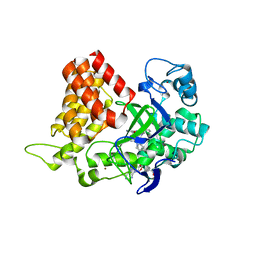 | | SMYD2 in complex with AZ931 | | Descriptor: | 6-[2-[4-[2-(3,4-dichlorophenyl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
4ZJC
 
 | | Structures of the human OX1 orexin receptor bound to selective and dual antagonists | | Descriptor: | OLEIC ACID, [5-(2-fluorophenyl)-2-methyl-1,3-thiazol-4-yl]{(2S)-2-[(5-phenyl-1,3,4-oxadiazol-2-yl)methyl]pyrrolidin-1-yl}methanone, human OX1R fusion protein to P.abysii glycogen synthase | | Authors: | Yin, J, Brautigam, C.A, Shao, Z, Clark, L, Harrell, C.M, Gotter, A.L, Coleman, P.J, Renger, J.J, Rosenbaum, D.M. | | Deposit date: | 2015-04-29 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.832 Å) | | Cite: | Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KSH
 
 | |
