8EW8
 
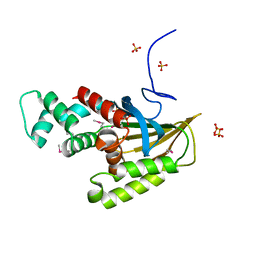 | | Crystal structure of Saccharomyces cerevisiae Altered Inheritance rate of Mitochondria protein 18 (AIM18p) R123A mutant | | Descriptor: | Altered inheritance of mitochondria protein 18, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Schmitz, J.M, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Aim18p and Aim46p are chalcone isomerase domain-containing mitochondrial hemoproteins in Saccharomyces cerevisiae.
J.Biol.Chem., 299, 2023
|
|
8EW9
 
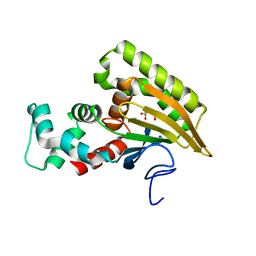 | | Crystal structure of Saccharomyces cerevisiae Altered Inheritance rate of Mitochondria protein 46 (AIM46p) | | Descriptor: | 2-OXOGLUTARIC ACID, Altered inheritance of mitochondria protein 46, mitochondrial | | Authors: | Bingman, C.A, Schmitz, J.M, Smith, R.W, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aim18p and Aim46p are chalcone isomerase domain-containing mitochondrial hemoproteins in Saccharomyces cerevisiae.
J.Biol.Chem., 299, 2023
|
|
6X6E
 
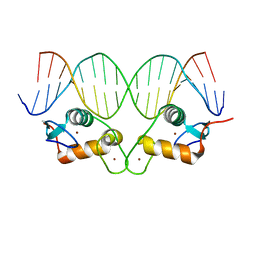 | |
8EPX
 
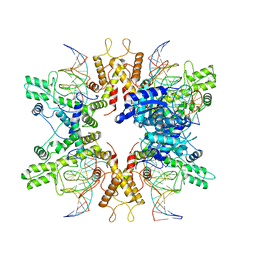 | | Type IIS Restriction Endonuclease PaqCI, DNA bound | | Descriptor: | CALCIUM ION, DNA 1a, DNA 1b, ... | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
7PUU
 
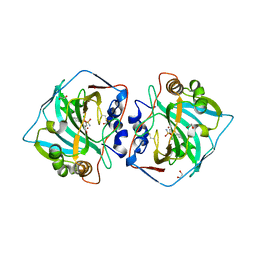 | | Crystal structure of carbonic anhydrase XII with methyl 4-chloro-2-cyclohexylsulfanyl-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0D
 
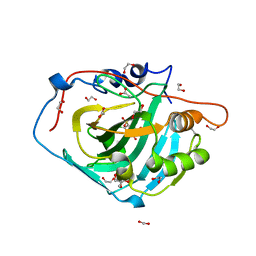 | | Human carbonic anhydrase I in complex with Methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0E
 
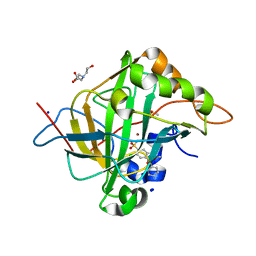 | | Human Carbonic Anhydrase II in complex with Methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | BICINE, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUW
 
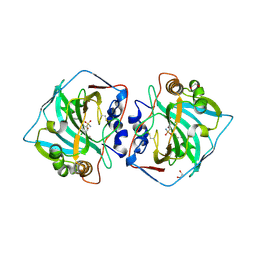 | | Crystal structure of carbonic anhydrase XII with methyl 2-chloro-4-[(2-phenylethyl)sulfanyl]-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
6X61
 
 | |
7PUV
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0C
 
 | | Mimic carbonic anhydrase IX in complex with Methyl 2-chloro-4-(cyclohexylsulfanyl)-5-sulfamoylbenzoate | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
6XI6
 
 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | Descriptor: | helical fusion design | | Authors: | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XH5
 
 | |
7PUO
 
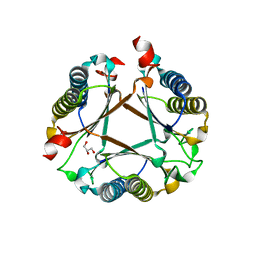 | | Structure of a fused 4-OT variant engineered for asymmetric Michael addition reactions | | Descriptor: | 2-hydroxymuconate tautomerase,Chains: A,B,C,D,E,F,2-hydroxymuconate tautomerase, CHLORIDE ION, GLYCEROL | | Authors: | Rozeboom, H.J, Thunnissen, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gene Fusion and Directed Evolution to Break Structural Symmetry and Boost Catalysis by an Oligomeric C-C Bond-Forming Enzyme.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6XAT
 
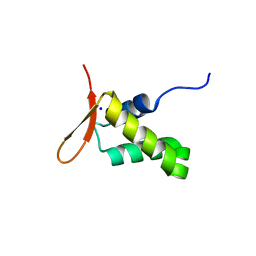 | | Crystal structure of the human FoxP4 DNA binding Domain | | Descriptor: | FOXP4 protein, SODIUM ION | | Authors: | VIllalobos, P, Castro-Fernandez, V, Medina, E, Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unraveling the folding and dimerization properties of the human FoxP subfamily of transcription factors.
Febs Lett., 597, 2023
|
|
6OC3
 
 | |
7NLH
 
 | |
6U8D
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
8EQN
 
 | |
8EUQ
 
 | | Crystal structure of HLA-DRA*01:01/HLA-DRB1*04:01 in complex with c44H10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Kassardjian, A, Julien, J.-P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Modular adjuvant-free pan-HLA-DR-immunotargeting subunit vaccine against SARS-CoV-2 elicits broad sarbecovirus-neutralizing antibody responses.
Cell Rep, 42, 2023
|
|
6XFR
 
 | |
8EYM
 
 | | CRYSTAL STRUCTURE OF NAGB-II PHOSPHOSUGAR ISOMERASE FROM SHEWANELLA DENITRIFICANS OS217 IN COMPLEX WITH GLUCITOLAMINE-6-PHOSPHATE AND N-ACETYLGLUCOSAMINE-6-PHOSPHATE AT 2.31 A RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rodriguez-Hernandez, A, Marcos-Viquez, J, Rodriguez-Romero, A, Bustos-Jaimes, I. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate binding in the allosteric site mimics homotropic cooperativity in the SIS-fold glucosamine-6-phosphate deaminases.
Protein Sci., 32, 2023
|
|
6S12
 
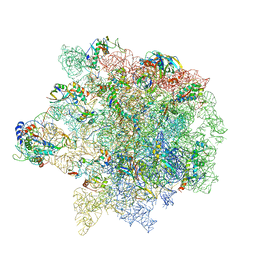 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22). | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
8EOL
 
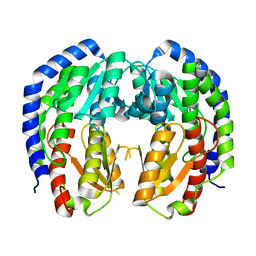 | |
6RW5
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF2 and IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
