4W9O
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6CFY
 
 | | Bosea sp Root 381 apo GapR structure | | Descriptor: | UPF0335 protein ASE63_04290 | | Authors: | Schumacherr, M.A. | | Deposit date: | 2018-02-18 | | Release date: | 2018-09-12 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
1JFN
 
 | | SOLUTION STRUCTURE OF HUMAN APOLIPOPROTEIN(A) KRINGLE IV TYPE 6 | | Descriptor: | APOLIPOPROTEIN A, KIV-T6 | | Authors: | Maderegger, B, Bermel, W, Hrzenjak, A, Kostner, G.M, Sterk, H. | | Deposit date: | 2001-06-21 | | Release date: | 2002-06-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human apolipoprotein(a) kringle IV type 6.
Biochemistry, 41, 2002
|
|
2VIR
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, LAMBDA), ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
3ZE8
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
8U9E
 
 | | Crystal Structure of Staphylococcus aureus Pdx1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Barra, A.L.C, Nascimento, A.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface
J.Biol.Chem., 300, 2024
|
|
5UYJ
 
 | | Crystal Structure of the Human CAMKK2B | | Descriptor: | 2-cyclopentyl-4-(7-methoxyquinolin-4-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, Drewry, D, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Human CAMKK2B
To Be Published
|
|
2VT4
 
 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND CYANOPINDOLOL | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, BETA1 ADRENERGIC RECEPTOR, DECANE, ... | | Authors: | Warne, A, Serrano-Vega, M.J, Baker, J.G, Moukhametzianov, R, Edwards, P.C, Henderson, R, Leslie, A.G.W, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2008-05-09 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Beta1-Adrenergic G-Protein-Coupled Receptor.
Nature, 454, 2008
|
|
1JLU
 
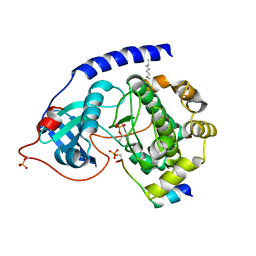 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
4IJH
 
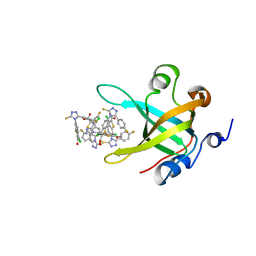 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | 3-chloro-6-[3-(4-fluorophenyl)-5-sulfanyl-4H-1,2,4-triazol-4-yl]-1-benzothiophene-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
1JO1
 
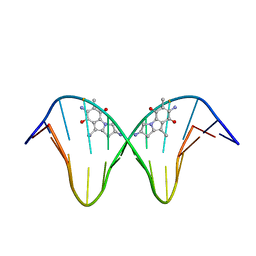 | | N7-Guanine Adduct of 2,7-diaminomitosene with DNA | | Descriptor: | 5'-D(*GP*TP*GP*(DAJ)GP*TP*AP*TP*AP*CP*CP*AP*C)-3', DECARBAMOYL-2,7-DIAMINOMITOSENE | | Authors: | Subramaniam, G, Paz, M.M, Kumar, G.S, Das, A, Palom, Y, Clement, C.C, Patel, D.J, Tomasz, M. | | Deposit date: | 2001-07-26 | | Release date: | 2001-09-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a guanine-N7-linked complex of the mitomycin C metabolite 2,7-diaminomitosene and DNA. Basis of sequence selectivity.
Biochemistry, 40, 2001
|
|
3X01
 
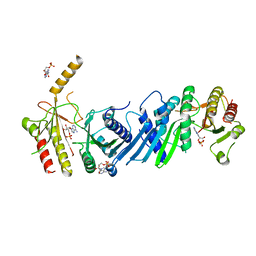 | | Crystal structure of PIP4KIIBETA complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
4V5T
 
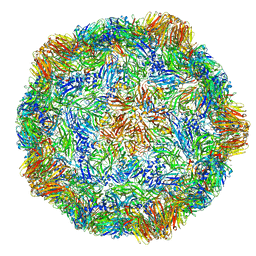 | | X-ray structure of the Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2011-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
7AM1
 
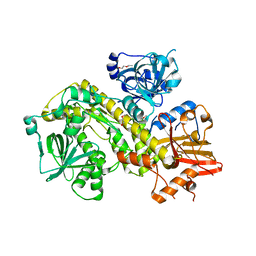 | | Structure of yeast Ssd1, a pseudonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protein SSD1 | | Authors: | Cook, A.G, Jayachandran, U. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast Ssd1 is a non-enzymatic member of the RNase II family with an alternative RNA recognition site.
Nucleic Acids Res., 50, 2022
|
|
4WCT
 
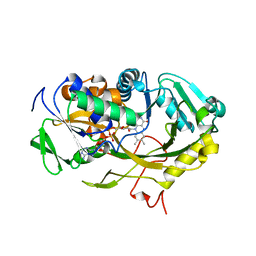 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
5D6D
 
 | | Crystal structure of GASDALIE IgG1 Fc in complex with FcgRIIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor III-A, ... | | Authors: | Ahmed, A.A, Bjorkman, P.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural characterization of GASDALIE Fc bound to the activating Fc receptor Fc gamma RIIIa.
J.Struct.Biol., 194, 2016
|
|
1JMZ
 
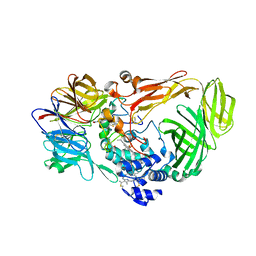 | | crystal structure of a quinohemoprotein amine dehydrogenase from pseudomonas putida with inhibitor | | Descriptor: | Amine Dehydrogenase, HEME C, NICKEL (II) ION, ... | | Authors: | Satoh, A, Miyahara, I, Hirotsu, K. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges.
J.Biol.Chem., 277, 2002
|
|
3XIS
 
 | |
6CQU
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase with Reactivator HI-6 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
2VUO
 
 | | Crystal structure of the rabbit IgG Fc fragment | | Descriptor: | AZIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Girardi, E, Holdom, M.D, Davies, A.M, Sutton, B.J, Beavil, A.J. | | Deposit date: | 2008-05-27 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Rabbit Igg-Fc.
Biochem.J., 417, 2009
|
|
6CJZ
 
 | |
8U7J
 
 | | Crystal Structure of Staphylococcus aureus PLP synthase complex | | Descriptor: | GLUTAMINE, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PdxS, ... | | Authors: | Barra, A.L.C, Brognaro, H, Betzel, C, Nascimento, A.S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface
J.Biol.Chem., 300, 2024
|
|
5V4Y
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-09510 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2017-03-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GRL-09510, a Unique P2-Crown-Tetrahydrofuranylurethane -Containing HIV-1 Protease Inhibitor, Maintains Its Favorable Antiviral Activity against Highly-Drug-Resistant HIV-1 Variants in vitro.
Sci Rep, 7, 2017
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
