7O43
 
 | |
7O3T
 
 | | I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3J
 
 | | O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein, TrwH protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-01 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O42
 
 | | TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwK protein,Protein hcp1 | | Authors: | Vadakkepat, A.K, Mace, K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
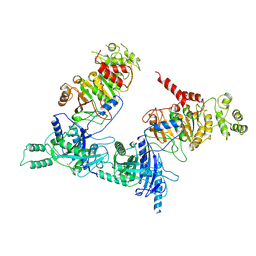
7NSK
 
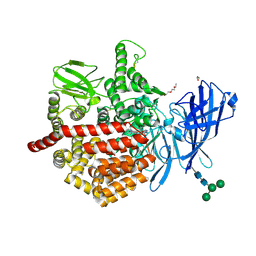 | | Endoplasmic reticulum aminopeptidase 2 complexed with a hydroxamic ligand | | Descriptor: | (2~{S})-3-(4-hydroxyphenyl)-~{N}-oxidanyl-2-[4-[[(5-pyridin-2-ylthiophen-2-yl)sulfonylamino]methyl]-1,2,3-triazol-1-yl]propanamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Mpakali, A, Giastas, P, Stratikos, E. | | Deposit date: | 2021-03-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Endoplasmic reticulum aminopeptidase 2 in complex with a phosphinic ligand
To Be Published
|
|
7OIU
 
 | | Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
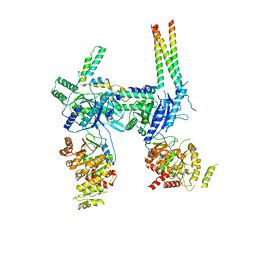
7O41
 
 | | Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O23
 
 | |
7QKN
 
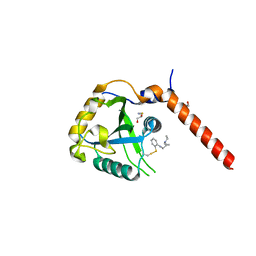 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 7 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8FWE
 
 | | Neck structure of Agrobacterium phage Milano, C3 symmetry | | Descriptor: | Collar sheath protein, gp13, Neck 1 protein, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8FYG
 
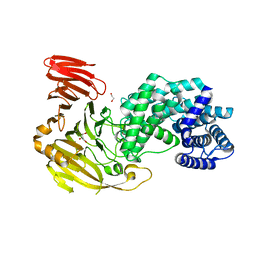 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
7NSN
 
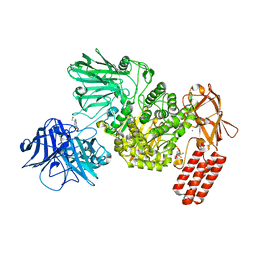 | | Multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis: mannoimidazole complex | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Kolaczkowski, B.M, Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S, Moeler, M.S, Meyer, A.S, Westh, P, Jensen, K, Krogh, K.B.R.M. | | Deposit date: | 2021-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and functional characterization of a multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6YUM
 
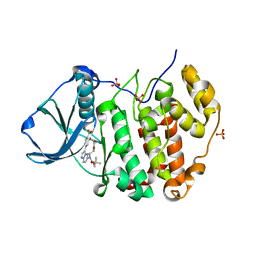 | | CK2 alpha bound to unclosed Macrocycle | | Descriptor: | 4-[5-[2-(2-hydroxyethyloxy)ethyl-[(2-methylpropan-2-yl)oxycarbonyl]amino]pyrazolo[1,5-a]pyrimidin-3-yl]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
5MWI
 
 | |
6YSO
 
 | | Crystal structure of the (SR) Ca2+-ATPase solved by vanadium SAD phasing | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
6YWZ
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 2-[(~{E})-(4-oxidanylidenebutanoylhydrazinylidene)methyl]benzoic acid, ARGININE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
7QHZ
 
 | | Crystal structure of KLK6 in complex with compound DKFZ917 | | Descriptor: | (5~{R})-3-(4-carbamimidoylphenyl)-~{N}-[(1~{S})-1-naphthalen-1-ylpropyl]-2-oxidanylidene-1,3-oxazolidine-5-carboxamide, GLYCEROL, Kallikrein-6 | | Authors: | Jagtap, P.K.A, Baumann, A, Lohbeck, J, Isak, D, Miller, A, Hennig, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Scalable synthesis and structural characterization of reversible KLK6 inhibitors.
Rsc Adv, 12, 2022
|
|
7QTV
 
 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7OFE
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 2-[(9-oxidanylidenefluoren-4-yl)carbonylamino]ethanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
8FRL
 
 | | Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and a macrocyclic peptide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, (7S,10S,13S,17P)-10-(4-aminobutyl)-7-(3-aminopropyl)-17-(6-aminopyridin-3-yl)-20-chloro-13-[(1H-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5H)-trione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
7OF8
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | (2~{S})-2-cyclopentyl-2-oxidanyl-2-phenyl-ethanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
8FRO
 
 | | Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and a macrocyclic peptide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, (7S,10S,13S)-10-(4-aminobutyl)-7-(3-aminopropyl)-17,20-dichloro-13-[(1H-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5H)-trione, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
7QQ0
 
 | |
7PAZ
 
 | | REDUCED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
7QEJ
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
