5SB9
 
 | | Tubulin-maytansinoid-4a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5W7J
 
 | | X-ray structure of the E89A variant of ankyrin repeat domain of DHHC17 in complex with Snap25b peptide | | Descriptor: | Palmitoyltransferase ZDHHC17, Snap25b-111-120 | | Authors: | Verardi, R, Kim, J.-S, Ghirlando, R, Banerjee, A. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.
Structure, 25, 2017
|
|
2YDZ
 
 | | X-ray structure of the cyan fluorescent protein SCFP3A (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
5F52
 
 | | Erwinia chrysanthemi L-asparaginase + Aspartic acid | | Descriptor: | ASPARTIC ACID, DI(HYDROXYETHYL)ETHER, L-asparaginase | | Authors: | Nguyen, H.A, Lavie, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Insight into Substrate Selectivity of Erwinia chrysanthemi l-Asparaginase.
Biochemistry, 55, 2016
|
|
4Q5N
 
 | |
6GWE
 
 | | Crystal structure of Thrombin bound to P2 macrocycle | | Descriptor: | (10S,14S,17R)-14-(3-carbamimidamidopropyl)-3-[[2-(hydroxymethyl)phenyl]methyl]-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(21),22,24-triene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cendron, L, Angelini, A, Kale, S.S, Bergeron-Brlek, M, Wu, Y, Heinis, C. | | Deposit date: | 2018-06-23 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thiol-to-amine cyclization reaction enables screening of large libraries of macrocyclic compounds and the generation of sub-kilodalton ligands.
Sci Adv, 5, 2019
|
|
6D7T
 
 | | Cryo-EM structure of human TRPV6-Y467A in complex with 2-Aminoethoxydiphenyl borate (2-APB) | | Descriptor: | 2-aminoethyl diphenylborinate, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
6Q2Z
 
 | | NMR solution structure of the HVO_2922 protein from Haloferax volcanii | | Descriptor: | UPF0339 family protein | | Authors: | Kubatova, N, Jonker, H.R.A, Saxena, K, Richter, C, Marchfelder, A, Schwalbe, H. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Small Protein HVO_2922 from Haloferax volcanii.
Chembiochem, 21, 2020
|
|
5AMP
 
 | | Geotrichum candidum Cel7A apo structure at 2.1A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE I, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
4WM8
 
 | | Crystal Structure of Human Enterovirus D68 | | Descriptor: | DECANOIC ACID, VP1, VP2, ... | | Authors: | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-10-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
5Z25
 
 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
6QET
 
 | | [41-82]Gga-AvBD11 | | Descriptor: | Gallinacin-11 | | Authors: | Meudal, H, Loth, K, Delmas, A.F, Landon, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and evolution ofGga-AvBD11, the archetype of the structural avian-double-beta-defensin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QEU
 
 | | Gga-AvBD11 (Avian beta-defensin 11 from Gallus gallus) | | Descriptor: | Gallinacin-11 | | Authors: | Meudal, H, Loth, K, Delmas, A.F, Landon, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and evolution ofGga-AvBD11, the archetype of the structural avian-double-beta-defensin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5Z37
 
 | |
5LR1
 
 | | CRYSTAL STRUCTURE OF HSP90 IN COMPLEX WITH A003498614A. | | Descriptor: | 4-chloranyl-7-[(4-chloranyl-3,5-dimethyl-pyridin-2-yl)methyl]pyrrolo[2,3-d]pyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Vallee, F, Dupuy, A. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | CRYSTAL STRUCTURE OF HSP90 IN COMPLEX WITH A003498614A.
To Be Published
|
|
5WE1
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5ARH
 
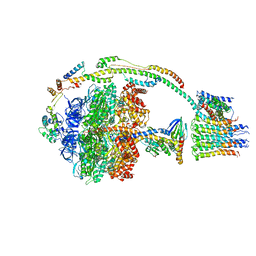 | | Bovine mitochondrial ATP synthase state 2a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
6Z0C
 
 | | Structure of in silico modelled artificial Maquette-3 protein | | Descriptor: | Maquette-3, POTASSIUM ION | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M.E, Asami, S, Mader, S.L, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of buried charged networks in artificial proteins
Nat Commun, 12, 2021
|
|
3KLK
 
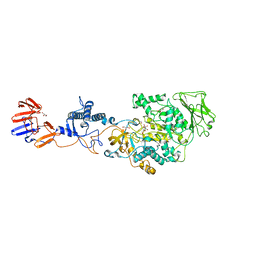 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4X25
 
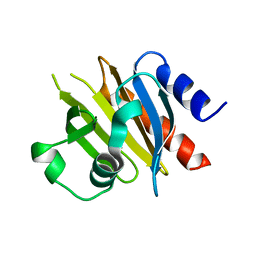 | |
4WW8
 
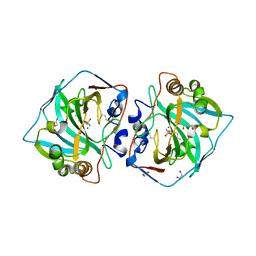 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-Propylthiobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-(propylsulfanyl)benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-11-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
3KFK
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (6.003 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
8CNF
 
 | |
6H2E
 
 | |
4Q9S
 
 | | Crystal Structure of human Focal Adhesion Kinase (Fak) bound to Compound1 (3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | Descriptor: | 3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, Focal adhesion kinase 1 | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
