4YW9
 
 | | Structure of rat cytosolic pepck in complex with 3-mercaptopicolinic acid and GTP | | Descriptor: | 3-sulfanylpyridine-2-carboxylic acid, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Balan, M.D, Johnson, T.A, Mcleod, M.J, Lotosky, W.R, Holyoak, T. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition and Allosteric Regulation of Monomeric Phosphoenolpyruvate Carboxykinase by 3-Mercaptopicolinic Acid.
Biochemistry, 54, 2015
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
3I5F
 
 | | Crystal structure of squid MG.ADP myosin S1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
8R8A
 
 | |
3FLM
 
 | | Crystal structure of menD from E.coli | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Priyadarshi, A, Hwang, K.Y. | | Deposit date: | 2008-12-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
5MKJ
 
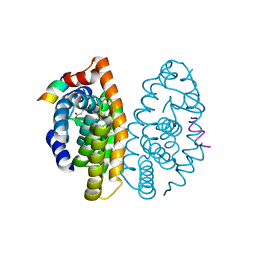 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 9 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(5-prop-2-enyl-2-propoxy-phenyl)phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
8R5C
 
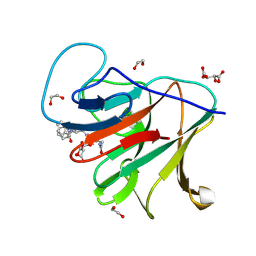 | | Crystal structure of human TRIM7 PRYSPRY domain bound to (2-(1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | (2~{S})-5-azanyl-5-oxidanylidene-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]pentanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
6N1G
 
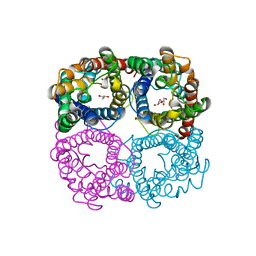 | |
5MLF
 
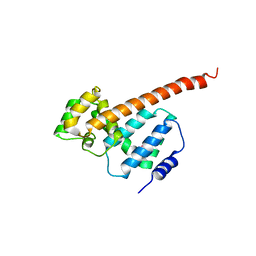 | | Structure of Psb29 at 1.55A | | Descriptor: | MERCURY (II) ION, Protein Thf1 | | Authors: | Murray, J.W, Kozlo, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.637 Å) | | Cite: | Structure of Psb29/Thf1 and its association with the FtsH protease complex involved in photosystem II repair in cyanobacteria.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
8R5D
 
 | | Crystal structure of human TRIM7 PRYSPRY domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, MALONIC ACID | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
6EO3
 
 | |
4RL9
 
 | |
6BXD
 
 | | Crystal structure of Variable Lymphocyte Receptor 2 (VLR2) | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Variable Lymphocyte Receptor 2 | | Authors: | Gunn, R.J, Wilson, I.A, Cooper, M.D, Herrin, B.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.103 Å) | | Cite: | VLR Recognition of TLR5 Expands the Molecular Characterization of Protein Antigen Binding by Non-Ig-based Antibodies.
J. Mol. Biol., 430, 2018
|
|
8F6E
 
 | | Cryo-EM structure of a Zinc-loaded wild-type YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab heavy chain, Fab light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
4MEP
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3-chloro-pyridone ligand | | Descriptor: | 3-chloro-5-[1-(3-methylpyridin-2-yl)-3-phenyl-1H-1,2,4-triazol-5-yl]pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
6EP6
 
 | | ARABIDOPSIS THALIANA GSTU23, reduced | | Descriptor: | GLYCEROL, Glutathione S-transferase U23, MAGNESIUM ION | | Authors: | Tossounian, M.A, Van Molle, I, Wahni, K, Jacques, S, Vertommen, D, Gevaert, K, Van Breusegem, F, Young, D, Rosado, L, Messens, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
6BY3
 
 | | Open and conductive conformation of KcsA-T75A mutant | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, DIACYL GLYCEROL, ... | | Authors: | Labro, A.J, Cortes, D.M, Tilegenova, C, Cuello, L.G. | | Deposit date: | 2017-12-19 | | Release date: | 2018-05-09 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Inverted allosteric coupling between activation and inactivation gates in K+channels.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2QM2
 
 | | Putative HopJ type III effector protein from Vibrio parahaemolyticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, POTASSIUM ION, ... | | Authors: | Kim, Y, Chang, C, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Putative HopJ type III Effector Protein from Vibrio parahaemolyticus.
To be Published
|
|
6EPG
 
 | |
1XVM
 
 | |
5T6I
 
 | | CRYSTAL STRUCTURE OF TGCDPK1 FROM TOXOPLASMA GONDII COMPLEXED WITH 5GB | | Descriptor: | 4-(3-methylphenyl)-5-(1,5-naphthyridin-2-yl)-1,3-thiazol-2-amine, Calmodulin-domain protein kinase 1 | | Authors: | Jiang, D.Q, Tempel, W, Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Osman, K.T, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of CDPK1 from toxoplasma gondii complexed with SGC-1-19
to be published
|
|
3H0C
 
 | | Crystal Structure of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-({(2S)-1-[(3R)-3-amino-4-(3-chlorophenyl)butanoyl]pyrrolidin-2-yl}methyl)-3-(methylsulfonyl)benzamide | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Deppe, H, Hill, O, Lopez-Canet, M, Rummey, C, Thiemann, M, Matassa, V.G, Edwards, P.J, Feurer, A. | | Deposit date: | 2009-04-09 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of b-homophenylalanine based pyrrolidin-2-ylmethyl amides and sulfonamides as highly potent and selective inhibitors of dipeptidyl peptidase IV
To be Published
|
|
8F6F
 
 | | Cryo-EM structure of a Zinc-loaded D51A mutant of the YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
6QFQ
 
 | | Structure of human Mcl-1 in complex with indole acid inhibitor | | Descriptor: | 7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
