4Z3M
 
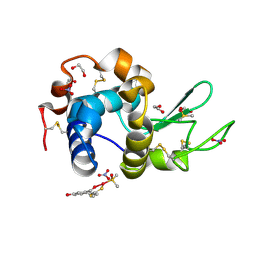 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-03-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
6WF6
 
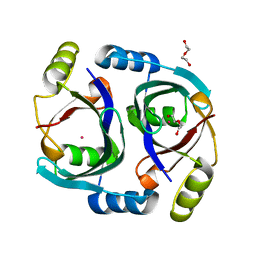 | | Streptomyces coelicolor methylmalonyl-CoA epimerase | | Descriptor: | COBALT (II) ION, DI(HYDROXYETHYL)ETHER, Methylmalonyl-CoA epimerase | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
6BKQ
 
 | |
8Q4U
 
 | |
7ZJS
 
 | |
6FRX
 
 | |
5YW2
 
 | |
8Q4Q
 
 | |
7ZLO
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 12 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
5JKN
 
 | | Crystal structure of deubiquitinase MINDY-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERCURY (II) ION, PHOSPHATE ION, ... | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes.
Mol.Cell, 63, 2016
|
|
8Q4R
 
 | |
5FIB
 
 | | Open form of murine Acid Sphingomyelinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
8Q4V
 
 | | Crystal structure of YTHDC1 in complex with Compound 37 (ZA_356) | | Descriptor: | 2-chloranyl-9-[(3-chlorophenyl)methyl]-~{N}-cyclopropyl-7,8-dihydropurin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Caflisch, A. | | Deposit date: | 2023-08-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
8QEV
 
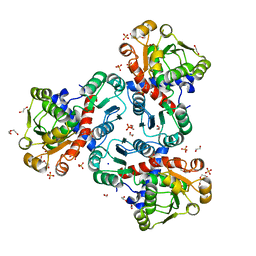 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
7ZLN
 
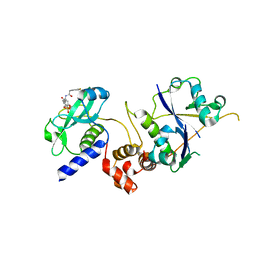 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 11 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
5FAS
 
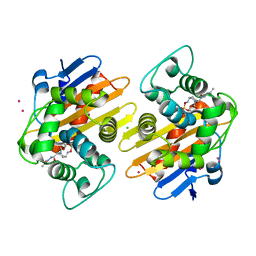 | | OXA-48 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
8Q4P
 
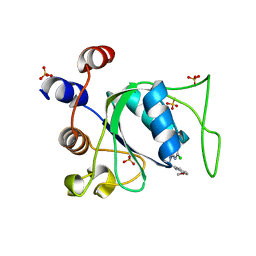 | |
6G56
 
 | |
8Q4W
 
 | | Crystal structure of YTHDC1 in complex with Compound 40 (CS_3a) | | Descriptor: | 5-chloranyl-3-[(3-chlorophenyl)methyl]-~{N}-methyl-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Caflisch, A. | | Deposit date: | 2023-08-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
8Q4N
 
 | |
8WZV
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 1 microsecond after photoexcitation with 40 microJoules laser intensity at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
4Z9G
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 in a hexagonal setting with translational non-crystallographic symmetry | | Descriptor: | 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, Corticotropin-releasing factor receptor 1,Lysozyme,Corticotropin-releasing factor receptor 1, OLEIC ACID, ... | | Authors: | Dore, A.S, Bortolato, A, Hollenstein, K, Cheng, R.K.Y, Read, R.J, Marshall, F.H. | | Deposit date: | 2015-04-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.183 Å) | | Cite: | Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures.
Curr Mol Pharmacol, 10, 2017
|
|
8Q4T
 
 | |
5FC1
 
 | | Murine SMPDL3A in complex with sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
6LKO
 
 | |
