7MB5
 
 | |
6BK7
 
 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
8UDT
 
 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
6VNA
 
 | | Pden_1323 | | Descriptor: | Pyridoxamine 5'-phosphate oxidase-related, FMN-binding protein | | Authors: | Isiorho, E.A, Mansoorabadi, S.O. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A noncanonical heme oxygenase specific for the degradation of c-type heme.
J.Biol.Chem., 296, 2021
|
|
6ZP1
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7MB6
 
 | |
5N29
 
 | |
8BIV
 
 | | Cystathionine gamma-lyase N360S mutant from Toxoplasma gondii | | Descriptor: | Cystathionine beta-lyase, putative, GLYCEROL, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
6P9V
 
 | | Crystal Structure of hMAT Mutant K289L | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Miller, M.D, Xu, W, Huber, T.D, Clinger, J.A, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.
Acs Chem.Biol., 15, 2020
|
|
5N5M
 
 | |
8BIX
 
 | | Cystathionine gamma-lyase N360S mutant from Toxoplasma gondii in complex with cystathionine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
6BZA
 
 | | Crystal structure of halogenase PltM in complex with phloroglucinol and FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Halogenase PltM, ... | | Authors: | Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-12-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unusual substrate and halide versatility of phenolic halogenase PltM.
Nat Commun, 10, 2019
|
|
4B7Y
 
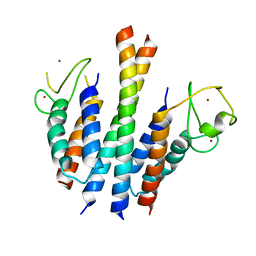 | | Crystal structure of the MSL1-MSL2 complex | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 2 HOMOLOG, ZINC ION | | Authors: | Hallacli, E, Lipp, M, Georgiev, P, Spielman, C, Cusack, S, Akhtar, A, Kadlec, J. | | Deposit date: | 2012-08-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Msl1-Mediated Dimerization of the Dosage Compensation Complex is Essential for Male X-Chromosome Regulation in Drosophila.
Mol.Cell, 48, 2012
|
|
6R5Z
 
 | |
6ZS2
 
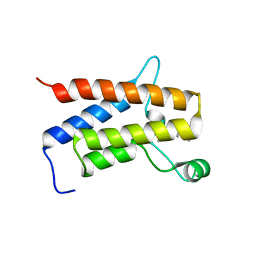 | | Crystal Structure of the bromodomain of human transcription activator BRG1 (SMARCA4) in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Transcription activator BRG1 | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6MQL
 
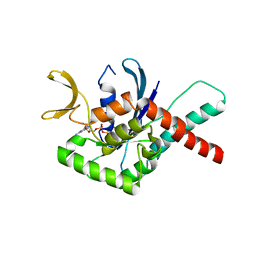 | | Crystal Structure of GTPase Domain of Human Septin 12 Mutant T89M | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-12 | | Authors: | Castro, D.K.S.V, Pereira, H.M, Brandao-Neto, J, Ulian, A.P.U, Garratt, R.C. | | Deposit date: | 2018-10-10 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of GTPase Domain of Human Septin 12
To Be Published
|
|
6GRK
 
 | |
5W7P
 
 | | Crystal structure of OxaC | | Descriptor: | OxaC, S-ADENOSYLMETHIONINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
3FVV
 
 | |
6GS7
 
 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6IVV
 
 | | Structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii with multiple surface binding regions at 1.26A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of peptide t-RNA hydrolase from Acinetobacter baumannii with multiple surface binding sites at 1.26 Angstrom resolution.
To Be Published
|
|
6ZVK
 
 | | The Halastavi arva virus (HalV) intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES17, ... | | Authors: | Abaeva, I.S, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
4PFJ
 
 | | The structure of bi-acetylated SAHH | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kavran, J.M, Wang, Y, Cole, P.A, Leahy, D.J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of s-adenosylhomocysteine hydrolase by lysine acetylation.
J.Biol.Chem., 289, 2014
|
|
6C4S
 
 | |
3M14
 
 | | Carbonic Anhydrase II in complex with novel sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, N-[(2Z)-1,3-thiazolidin-2-ylidene]sulfamide, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G, Sandner, N.U. | | Deposit date: | 2010-03-04 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural investigation and inhibitor studies on Carbonic Anhydrase II
To be Published
|
|
