6QSF
 
 | | Crystal structure of Pizza6S in the presence of Keggin (STA) | | Descriptor: | Keggin (STA), Pizza6S | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
8QA1
 
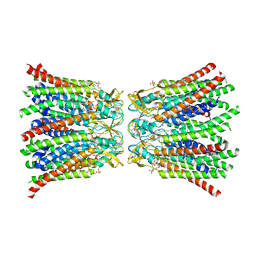 | |
7JVA
 
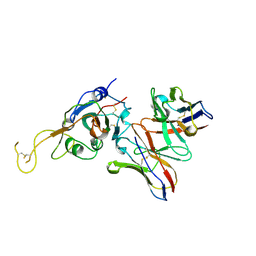 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | Descriptor: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6G59
 
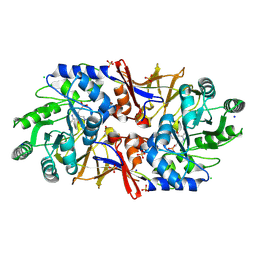 | | Structure of the alanine racemase from Staphylococcus aureus in complex with an pyridoxal-6- phosphate derivative | | Descriptor: | (6-ethynyl-4-methanoyl-5-oxidanyl-pyridin-3-yl)methyl dihydrogen phosphate, Alanine racemase 1, CHLORIDE ION, ... | | Authors: | Hoegl, A, Sieber, S.A, Schneider, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mining the cellular inventory of pyridoxal phosphate-dependent enzymes with functionalized cofactor mimics.
Nat Chem, 10, 2018
|
|
8Q9Z
 
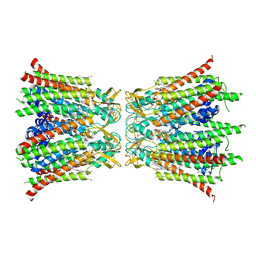 | |
8GHZ
 
 | | Cryo-EM structure of fish immunogloblin M-Fc | | Descriptor: | Teleost immunoglobulin M protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lyu, M, Stadtmueller, B.M, Malyutin, A.G. | | Deposit date: | 2023-03-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The structure of the teleost Immunoglobulin M core provides insights on polymeric antibody evolution, assembly, and function.
Nat Commun, 14, 2023
|
|
3N3A
 
 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli in Complex with Reduced NrdI | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein nrdI, ... | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
6G60
 
 | |
8QA0
 
 | |
6NF0
 
 | | Nocturnin with bound NADPH substrate | | Descriptor: | CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nocturnin | | Authors: | Estrella, M.A, Du, J, Korennykh, A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The metabolites NADP+and NADPH are the targets of the circadian protein Nocturnin (Curled).
Nat Commun, 10, 2019
|
|
6RAJ
 
 | | Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6G74
 
 | |
7SFQ
 
 | | EmrE S64V Mutant Bound to tetra(4-fluorophenyl)phosphonium at pH 8.0 | | Descriptor: | Multidrug transporter EmrE, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Spreacker, P.J, Dregni, A.J, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2021-10-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport.
Nat Commun, 13, 2022
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
8QWL
 
 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QVV
 
 | | Crystal structure of Ompk36 GD at 3500 eV based on analytical absorption corrections | | Descriptor: | OmpK36, SULFATE ION | | Authors: | Duman, R, Wagner, A, Beis, K, Wong, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
4TF4
 
 | | ENDO/EXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-31 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
5FY5
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with fumarate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Fumarate
To be Published
|
|
4ZDX
 
 | | Structure of OXA-51 beta-lactamase | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Beta-lactamase, GLYCEROL | | Authors: | Smith, C.A, Antunes, N.T, Stewart, N.K, Frase, H, Toth, M, Kantardjieff, K.A, Vakulenko, S.B. | | Deposit date: | 2015-04-20 | | Release date: | 2015-06-17 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Enhancement of Carbapenemase Activity in the OXA-51 Family of Class D beta-Lactamases.
Acs Chem.Biol., 10, 2015
|
|
5FYI
 
 | | Crystal structure of human JMJD2A in complex with pyruvate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Szykowska, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal Structure of Human Jmjd2A in Complex with Pyruvate
To be Published
|
|
6RAK
 
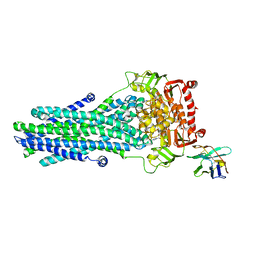 | | Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
8Q3C
 
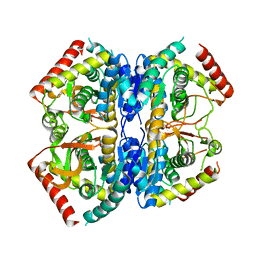 | | Structure of Selenomonas ruminantium lactate dehydrogenase I85R mutant | | Descriptor: | CHLORIDE ION, L-lactate dehydrogenase, NITRATE ION, ... | | Authors: | Bertrand, Q, Coquille, S, Iorio, A, Sterpone, F, Madern, D. | | Deposit date: | 2023-08-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
5O3S
 
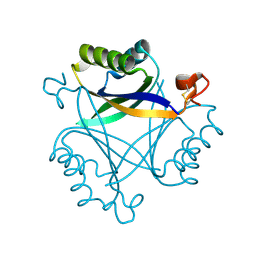 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in Apo state, hexagonal crystal form | | Descriptor: | Membrane-associated protein slr1513 | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2017-05-24 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PII-like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FPZ
 
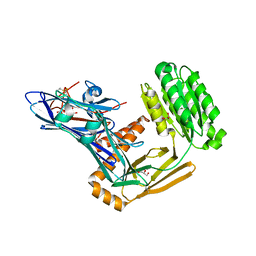 | | Inter-alpha-inhibitor heavy chain 1, D298A | | Descriptor: | ACETATE ION, GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1 | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
4Z6A
 
 | | Crystal Structure of a FVIIa-Trypsin Chimera (YT) in Complex with Soluble Tissue Factor | | Descriptor: | CALCIUM ION, CITRIC ACID, Coagulation factor VII, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2015-04-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity.
J.Biol.Chem., 291, 2016
|
|
