7QWH
 
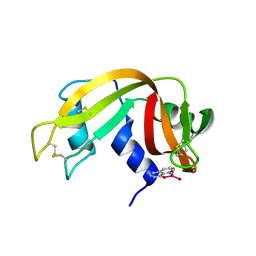 | | X-ray structure of the adduct formed upon reaction of a vanadium hydroxyquinoline complex with RNase A | | Descriptor: | 2,2-bis($l^{1}-oxidanyl)-3-oxa-1$l^{4}-aza-2$l^{4}-vanadatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaene, Ribonuclease pancreatic | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Interaction of VIVO-8-hydroxyquinoline species with RNase A: the effect of metal ligands in the protein adduct stabilization
Inorg Chem Front, 2023
|
|
6UHK
 
 | | Crystal Structure of C176 mGFP | | Descriptor: | C176 mGFP | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6E1P
 
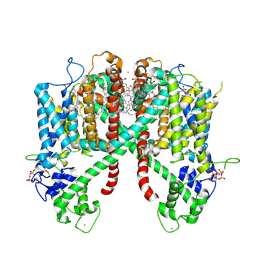 | | Structure of AtTPC1(DDE) in state 2 | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6OGR
 
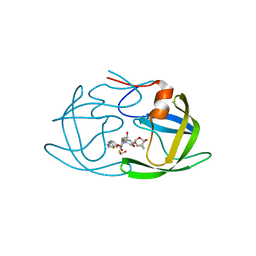 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
6RK4
 
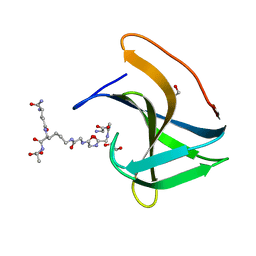 | | Lysostaphin SH3b P4-G5 complex, synchrotron dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, 1,2-ETHANEDIOL, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
5K78
 
 | | Dbr1 in complex with 16-mer branched RNA | | Descriptor: | FE (II) ION, RNA lariat debranching enzyme, putative, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
6RJ6
 
 | | Crystal structure of PHGDH in complex with BI-4924 | | Descriptor: | 2-[4-[(1~{S})-1-[[4,5-bis(chloranyl)-1,6-dimethyl-indol-2-yl]carbonylamino]-2-oxidanyl-ethyl]phenyl]sulfonylethanoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
7LIF
 
 | |
6HNB
 
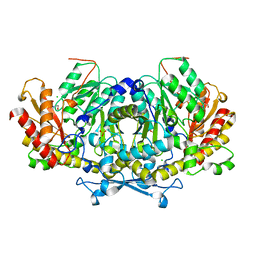 | | Crystal structure of aminotransferase Aro8 from Candida albicans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aromatic amino acid aminotransferase I, CHLORIDE ION, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
6HOI
 
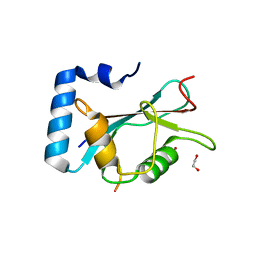 | | Structure of Beclin1 LIR motif bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beclin-1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
4QUH
 
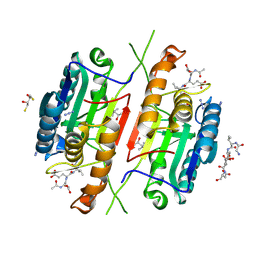 | | Caspase-3 T140G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
6HNL
 
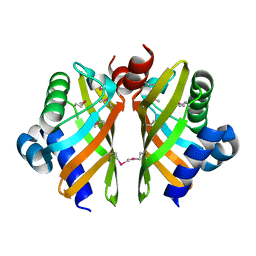 | | Selenomethionine derivative of IdmH 96-104 loop truncation variant | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6HNV
 
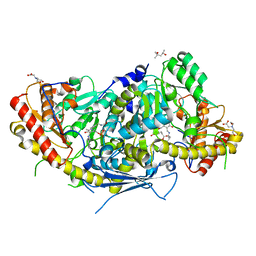 | | Crystal structure of aminotransferase Aro9 from C. Albicans with ligands | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINOHEXANEDIOIC ACID, 2-OXOADIPIC ACID, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
5JSX
 
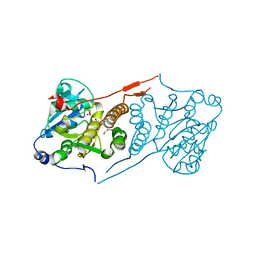 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+ and uridine-diphosphate-glucose (UDP-Glc) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
5JT4
 
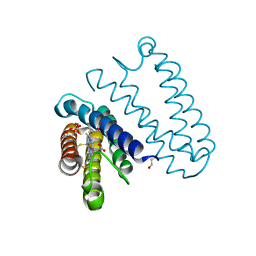 | | L16A mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous state | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
6HPQ
 
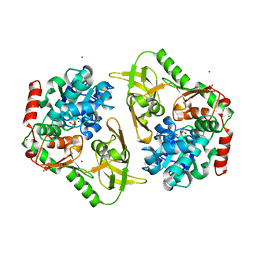 | | Crystal structure of human Pif1 helicase in complex with AMP-PNP, brominated crystal form. | | Descriptor: | ATP-dependent DNA helicase PIF1, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Ledikov, V.M, Dehghani-Tafti, S, Bax, B, Sanders, C.M, Antson, A.A. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Nucleic Acids Res., 47, 2019
|
|
5JTC
 
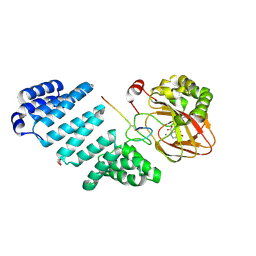 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH)oxygenase and TPR domains in complex with manganese, 2,4-pyridine dicarboxylate and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, BROMIDE ION, Coagulation factor X, ... | | Authors: | McDonough, M.A, Pfeffer, I. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Aspartate/asparagine-beta-hydroxylase: a high-throughput mass spectrometric assay for discovery of small molecule inhibitors.
Sci Rep, 10, 2020
|
|
6EJJ
 
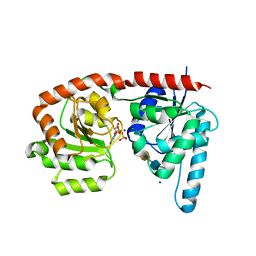 | | Structure of a glycosyltransferase / state 2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, NerylNeryl pyrophosphate, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
6EK6
 
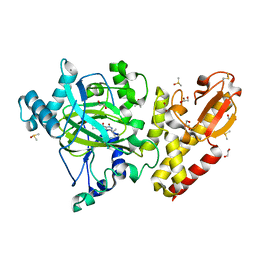 | | Crystal structure of KDM5B in complex with S49195a. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Srikannathasan, V, Szykowska, A, Newman, J.A, Ruda, G.F, Strain-Damerell, C, Burgess-Brown, N.A, Vazquez-Rodriguez, S, Wright, M, Brennan, P.E, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of KDM5B in complex with S49195a.
To be published
|
|
5KCW
 
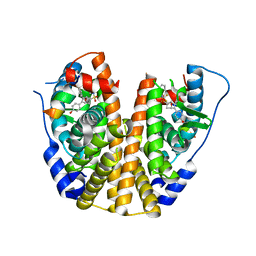 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-trifluoroethyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-phenyl-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
6HRD
 
 | |
5KD9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-trifluoroethyl 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(4-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5GG7
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP, 8-oxo-dGMP and pyrophosphate (I) | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGC
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with phosphate and magnesium ions (excess magnesium, I) | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
